
Tortoise microvirus 59
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.42
Get precalculated fractions of proteins
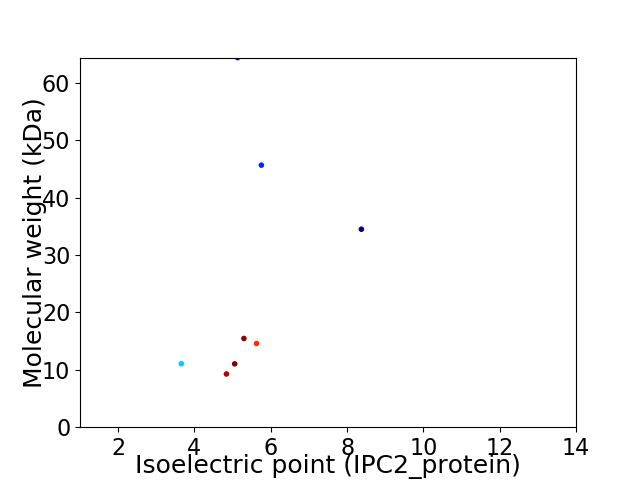
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
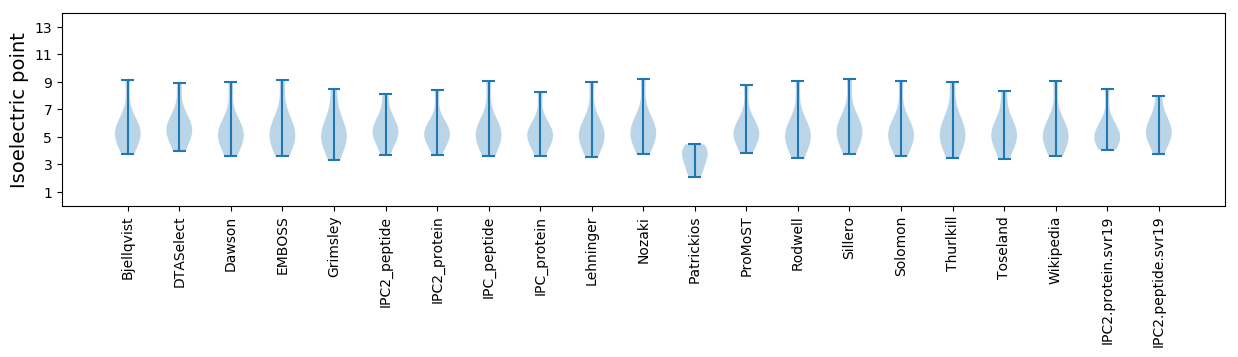
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVZ6|A0A4V1FVZ6_9VIRU Major capsid protein OS=Tortoise microvirus 59 OX=2583164 PE=3 SV=1
MM1 pKa = 7.76SDD3 pKa = 4.67FIPIDD8 pKa = 3.67FFTDD12 pKa = 3.02DD13 pKa = 4.25GYY15 pKa = 11.87APVPVRR21 pKa = 11.84YY22 pKa = 8.71LASPEE27 pKa = 3.86TAEE30 pKa = 3.88VFNRR34 pKa = 11.84VMSMAPTFFPDD45 pKa = 4.22LPVDD49 pKa = 4.28ANPVIAEE56 pKa = 3.94QQYY59 pKa = 11.45AMALVTLAVNFYY71 pKa = 10.73FDD73 pKa = 4.57ALGLLSPPEE82 pKa = 4.25VAPSTPVGDD91 pKa = 3.93DD92 pKa = 3.19VLDD95 pKa = 3.77IKK97 pKa = 10.49EE98 pKa = 4.2SRR100 pKa = 11.84VV101 pKa = 3.25
MM1 pKa = 7.76SDD3 pKa = 4.67FIPIDD8 pKa = 3.67FFTDD12 pKa = 3.02DD13 pKa = 4.25GYY15 pKa = 11.87APVPVRR21 pKa = 11.84YY22 pKa = 8.71LASPEE27 pKa = 3.86TAEE30 pKa = 3.88VFNRR34 pKa = 11.84VMSMAPTFFPDD45 pKa = 4.22LPVDD49 pKa = 4.28ANPVIAEE56 pKa = 3.94QQYY59 pKa = 11.45AMALVTLAVNFYY71 pKa = 10.73FDD73 pKa = 4.57ALGLLSPPEE82 pKa = 4.25VAPSTPVGDD91 pKa = 3.93DD92 pKa = 3.19VLDD95 pKa = 3.77IKK97 pKa = 10.49EE98 pKa = 4.2SRR100 pKa = 11.84VV101 pKa = 3.25
Molecular weight: 11.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6T5|A0A4P8W6T5_9VIRU Uncharacterized protein OS=Tortoise microvirus 59 OX=2583164 PE=4 SV=1
MM1 pKa = 7.48SCLSPVTIHH10 pKa = 6.46VNKK13 pKa = 10.0QSYY16 pKa = 7.94EE17 pKa = 4.27VPCRR21 pKa = 11.84QCLSCRR27 pKa = 11.84LDD29 pKa = 3.28YY30 pKa = 11.34QMYY33 pKa = 8.78LTFCVEE39 pKa = 3.77NEE41 pKa = 4.22LYY43 pKa = 10.68DD44 pKa = 3.74LYY46 pKa = 11.42KK47 pKa = 10.53RR48 pKa = 11.84GLGATFCTFTYY59 pKa = 10.88SDD61 pKa = 3.91VNLPSNGSLCKK72 pKa = 9.96SDD74 pKa = 3.76LQNLFKK80 pKa = 10.29TVRR83 pKa = 11.84TTAKK87 pKa = 10.17RR88 pKa = 11.84NGSLPPFKK96 pKa = 10.79YY97 pKa = 9.65IACGEE102 pKa = 4.18YY103 pKa = 10.04GDD105 pKa = 4.41KK106 pKa = 10.53FGRR109 pKa = 11.84PHH111 pKa = 5.0YY112 pKa = 10.21HH113 pKa = 6.23AVILGWSDD121 pKa = 3.85VICHH125 pKa = 5.67NMFRR129 pKa = 11.84KK130 pKa = 9.75YY131 pKa = 6.64WTHH134 pKa = 5.81GLSDD138 pKa = 4.08FGVLRR143 pKa = 11.84SGGVRR148 pKa = 11.84YY149 pKa = 9.49VLKK152 pKa = 10.73YY153 pKa = 7.99CTKK156 pKa = 10.14SVRR159 pKa = 11.84GTKK162 pKa = 10.19AEE164 pKa = 3.98EE165 pKa = 4.25MYY167 pKa = 10.64DD168 pKa = 3.53AQGLEE173 pKa = 4.27RR174 pKa = 11.84PFITHH179 pKa = 4.96SHH181 pKa = 6.54KK182 pKa = 10.43IGYY185 pKa = 8.85DD186 pKa = 3.31WLCRR190 pKa = 11.84NADD193 pKa = 4.24DD194 pKa = 4.84IVANNFTYY202 pKa = 10.66LNKK205 pKa = 10.15GVRR208 pKa = 11.84RR209 pKa = 11.84PIPKK213 pKa = 9.86YY214 pKa = 10.43FRR216 pKa = 11.84DD217 pKa = 3.46KK218 pKa = 11.04YY219 pKa = 11.44DD220 pKa = 3.2YY221 pKa = 10.93FKK223 pKa = 10.77TFNPMPAVDD232 pKa = 4.13FLRR235 pKa = 11.84RR236 pKa = 11.84EE237 pKa = 4.48GALHH241 pKa = 6.88NYY243 pKa = 8.63PDD245 pKa = 3.52YY246 pKa = 11.37RR247 pKa = 11.84EE248 pKa = 3.9WSKK251 pKa = 11.37VKK253 pKa = 10.42AYY255 pKa = 10.57NKK257 pKa = 10.13EE258 pKa = 3.58KK259 pKa = 11.02GLIQRR264 pKa = 11.84ARR266 pKa = 11.84QEE268 pKa = 4.66GIPVDD273 pKa = 3.93DD274 pKa = 5.32ANFIATPLPSPSLAPSLVTEE294 pKa = 4.22VLNRR298 pKa = 11.84PP299 pKa = 3.68
MM1 pKa = 7.48SCLSPVTIHH10 pKa = 6.46VNKK13 pKa = 10.0QSYY16 pKa = 7.94EE17 pKa = 4.27VPCRR21 pKa = 11.84QCLSCRR27 pKa = 11.84LDD29 pKa = 3.28YY30 pKa = 11.34QMYY33 pKa = 8.78LTFCVEE39 pKa = 3.77NEE41 pKa = 4.22LYY43 pKa = 10.68DD44 pKa = 3.74LYY46 pKa = 11.42KK47 pKa = 10.53RR48 pKa = 11.84GLGATFCTFTYY59 pKa = 10.88SDD61 pKa = 3.91VNLPSNGSLCKK72 pKa = 9.96SDD74 pKa = 3.76LQNLFKK80 pKa = 10.29TVRR83 pKa = 11.84TTAKK87 pKa = 10.17RR88 pKa = 11.84NGSLPPFKK96 pKa = 10.79YY97 pKa = 9.65IACGEE102 pKa = 4.18YY103 pKa = 10.04GDD105 pKa = 4.41KK106 pKa = 10.53FGRR109 pKa = 11.84PHH111 pKa = 5.0YY112 pKa = 10.21HH113 pKa = 6.23AVILGWSDD121 pKa = 3.85VICHH125 pKa = 5.67NMFRR129 pKa = 11.84KK130 pKa = 9.75YY131 pKa = 6.64WTHH134 pKa = 5.81GLSDD138 pKa = 4.08FGVLRR143 pKa = 11.84SGGVRR148 pKa = 11.84YY149 pKa = 9.49VLKK152 pKa = 10.73YY153 pKa = 7.99CTKK156 pKa = 10.14SVRR159 pKa = 11.84GTKK162 pKa = 10.19AEE164 pKa = 3.98EE165 pKa = 4.25MYY167 pKa = 10.64DD168 pKa = 3.53AQGLEE173 pKa = 4.27RR174 pKa = 11.84PFITHH179 pKa = 4.96SHH181 pKa = 6.54KK182 pKa = 10.43IGYY185 pKa = 8.85DD186 pKa = 3.31WLCRR190 pKa = 11.84NADD193 pKa = 4.24DD194 pKa = 4.84IVANNFTYY202 pKa = 10.66LNKK205 pKa = 10.15GVRR208 pKa = 11.84RR209 pKa = 11.84PIPKK213 pKa = 9.86YY214 pKa = 10.43FRR216 pKa = 11.84DD217 pKa = 3.46KK218 pKa = 11.04YY219 pKa = 11.44DD220 pKa = 3.2YY221 pKa = 10.93FKK223 pKa = 10.77TFNPMPAVDD232 pKa = 4.13FLRR235 pKa = 11.84RR236 pKa = 11.84EE237 pKa = 4.48GALHH241 pKa = 6.88NYY243 pKa = 8.63PDD245 pKa = 3.52YY246 pKa = 11.37RR247 pKa = 11.84EE248 pKa = 3.9WSKK251 pKa = 11.37VKK253 pKa = 10.42AYY255 pKa = 10.57NKK257 pKa = 10.13EE258 pKa = 3.58KK259 pKa = 11.02GLIQRR264 pKa = 11.84ARR266 pKa = 11.84QEE268 pKa = 4.66GIPVDD273 pKa = 3.93DD274 pKa = 5.32ANFIATPLPSPSLAPSLVTEE294 pKa = 4.22VLNRR298 pKa = 11.84PP299 pKa = 3.68
Molecular weight: 34.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1846 |
83 |
579 |
230.8 |
25.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.909 ± 1.287 | 1.517 ± 0.661 |
6.176 ± 0.484 | 4.55 ± 0.706 |
4.55 ± 0.544 | 5.796 ± 0.671 |
1.733 ± 0.257 | 4.28 ± 0.511 |
3.684 ± 0.989 | 9.534 ± 0.928 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.492 ± 0.267 | 4.984 ± 0.431 |
5.85 ± 0.767 | 4.659 ± 1.714 |
5.525 ± 0.427 | 8.667 ± 0.925 |
6.23 ± 0.545 | 6.176 ± 0.778 |
0.65 ± 0.157 | 5.038 ± 0.798 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
