
Torque teno virus 8
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
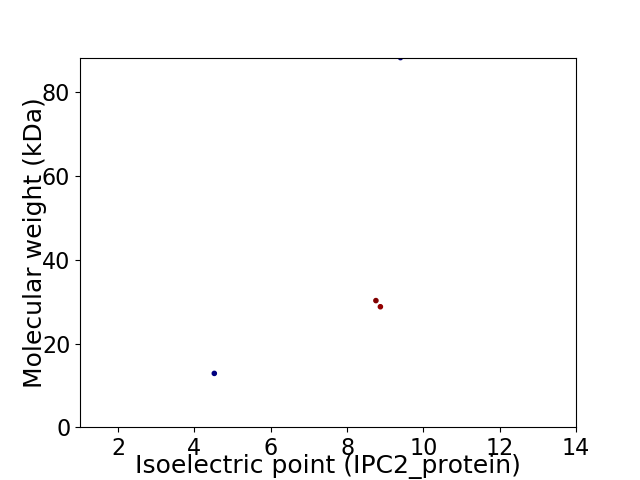
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q91PS9|Q91PS9_9VIRU Uncharacterized protein OS=Torque teno virus 8 OX=687347 PE=4 SV=1
MM1 pKa = 7.63SDD3 pKa = 4.45WIPPAQNVADD13 pKa = 4.96RR14 pKa = 11.84EE15 pKa = 4.47SDD17 pKa = 2.99WFRR20 pKa = 11.84AVYY23 pKa = 10.08CSHH26 pKa = 7.22KK27 pKa = 10.19CFCGCGDD34 pKa = 3.96PVRR37 pKa = 11.84HH38 pKa = 6.09LASISAAVGYY48 pKa = 8.72QPPPGSRR55 pKa = 11.84QAPGSSGTPPLIRR68 pKa = 11.84GLRR71 pKa = 11.84ALPAAPSGSSNQQCGGTAGRR91 pKa = 11.84GTGGDD96 pKa = 3.12RR97 pKa = 11.84SEE99 pKa = 4.66GGEE102 pKa = 3.88PTAAEE107 pKa = 4.28DD108 pKa = 4.78PYY110 pKa = 11.66DD111 pKa = 4.31GDD113 pKa = 4.77AEE115 pKa = 4.14EE116 pKa = 5.28LLAALEE122 pKa = 4.25DD123 pKa = 4.13VEE125 pKa = 4.66
MM1 pKa = 7.63SDD3 pKa = 4.45WIPPAQNVADD13 pKa = 4.96RR14 pKa = 11.84EE15 pKa = 4.47SDD17 pKa = 2.99WFRR20 pKa = 11.84AVYY23 pKa = 10.08CSHH26 pKa = 7.22KK27 pKa = 10.19CFCGCGDD34 pKa = 3.96PVRR37 pKa = 11.84HH38 pKa = 6.09LASISAAVGYY48 pKa = 8.72QPPPGSRR55 pKa = 11.84QAPGSSGTPPLIRR68 pKa = 11.84GLRR71 pKa = 11.84ALPAAPSGSSNQQCGGTAGRR91 pKa = 11.84GTGGDD96 pKa = 3.12RR97 pKa = 11.84SEE99 pKa = 4.66GGEE102 pKa = 3.88PTAAEE107 pKa = 4.28DD108 pKa = 4.78PYY110 pKa = 11.66DD111 pKa = 4.31GDD113 pKa = 4.77AEE115 pKa = 4.14EE116 pKa = 5.28LLAALEE122 pKa = 4.25DD123 pKa = 4.13VEE125 pKa = 4.66
Molecular weight: 12.86 kDa
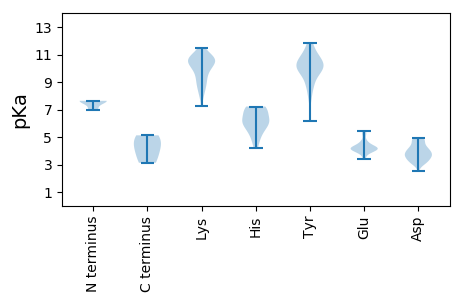
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91PS8|Q91PS8_9VIRU Uncharacterized protein OS=Torque teno virus 8 OX=687347 PE=4 SV=1
MM1 pKa = 7.0WWNRR5 pKa = 11.84WKK7 pKa = 10.57RR8 pKa = 11.84YY9 pKa = 6.18RR10 pKa = 11.84WRR12 pKa = 11.84PLRR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84TYY20 pKa = 9.03RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PLRR27 pKa = 11.84WRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84GAARR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84VRR43 pKa = 11.84RR44 pKa = 11.84WRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AWGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84YY57 pKa = 9.38IRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84LRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.74KK69 pKa = 10.57IPITQWNPSVVRR81 pKa = 11.84KK82 pKa = 9.76CVVTGYY88 pKa = 10.29IPLLICGTGTTGTTYY103 pKa = 11.21KK104 pKa = 10.81NYY106 pKa = 10.12GSHH109 pKa = 6.29AADD112 pKa = 3.47YY113 pKa = 10.8KK114 pKa = 11.06KK115 pKa = 10.68YY116 pKa = 10.53DD117 pKa = 3.87PFGGGYY123 pKa = 7.98STMQFTLEE131 pKa = 4.07TLFDD135 pKa = 3.37EE136 pKa = 4.96WVKK139 pKa = 10.68HH140 pKa = 4.8RR141 pKa = 11.84CTWSRR146 pKa = 11.84SNKK149 pKa = 9.64DD150 pKa = 3.42LEE152 pKa = 4.23LVRR155 pKa = 11.84YY156 pKa = 9.79LGGSFTLYY164 pKa = 10.2RR165 pKa = 11.84HH166 pKa = 6.36PKK168 pKa = 8.45CDD170 pKa = 4.42FIFRR174 pKa = 11.84YY175 pKa = 9.0NRR177 pKa = 11.84KK178 pKa = 8.92PPFKK182 pKa = 9.28DD183 pKa = 3.34TQITGPSLHH192 pKa = 7.02PGILMTRR199 pKa = 11.84RR200 pKa = 11.84KK201 pKa = 9.98KK202 pKa = 10.72KK203 pKa = 9.68IIKK206 pKa = 10.22SFLTRR211 pKa = 11.84PRR213 pKa = 11.84GRR215 pKa = 11.84ATKK218 pKa = 10.09RR219 pKa = 11.84VRR221 pKa = 11.84FKK223 pKa = 11.11PPTLYY228 pKa = 9.54TDD230 pKa = 2.56KK231 pKa = 10.66WYY233 pKa = 9.67FQKK236 pKa = 10.7DD237 pKa = 3.24ICNLPLVNIAASACSLRR254 pKa = 11.84FPFCSPQTDD263 pKa = 3.56NTCVNFQVLSKK274 pKa = 10.62RR275 pKa = 11.84FNTMLSISPDD285 pKa = 3.77YY286 pKa = 10.34PKK288 pKa = 10.91KK289 pKa = 10.8NYY291 pKa = 10.4DD292 pKa = 3.85SFVKK296 pKa = 10.0TYY298 pKa = 10.81LKK300 pKa = 10.43NAQEE304 pKa = 4.19HH305 pKa = 5.36WNQNKK310 pKa = 10.23KK311 pKa = 8.73GTLDD315 pKa = 3.54NYY317 pKa = 10.92NDD319 pKa = 4.12PNRR322 pKa = 11.84LLTVFNTFKK331 pKa = 10.63TEE333 pKa = 3.42EE334 pKa = 4.08HH335 pKa = 6.84LYY337 pKa = 10.89DD338 pKa = 4.33PSAKK342 pKa = 8.26EE343 pKa = 4.21TKK345 pKa = 9.14NTTNTDD351 pKa = 3.27AAGNKK356 pKa = 9.07YY357 pKa = 8.63NTVTSLWGDD366 pKa = 3.6YY367 pKa = 9.88IYY369 pKa = 11.16KK370 pKa = 8.65EE371 pKa = 4.21TVVDD375 pKa = 3.83NFINNAKK382 pKa = 10.47NYY384 pKa = 9.58FEE386 pKa = 4.21SRR388 pKa = 11.84KK389 pKa = 10.5GNIVMSNEE397 pKa = 3.84YY398 pKa = 10.46LNHH401 pKa = 5.55KK402 pKa = 8.56TGLYY406 pKa = 10.5SPIFLSNSRR415 pKa = 11.84LSPDD419 pKa = 3.13FPGFYY424 pKa = 9.38IDD426 pKa = 4.78VLYY429 pKa = 11.32NPANDD434 pKa = 3.59KK435 pKa = 11.5GIGNKK440 pKa = 7.93IWMDD444 pKa = 3.17WCTKK448 pKa = 10.56NDD450 pKa = 3.7STWRR454 pKa = 11.84DD455 pKa = 3.6TPQKK459 pKa = 10.46LPVVDD464 pKa = 3.74IPLWAALLGYY474 pKa = 10.11SDD476 pKa = 4.93YY477 pKa = 9.93CTKK480 pKa = 10.7YY481 pKa = 10.83FNDD484 pKa = 3.38KK485 pKa = 11.13GLIKK489 pKa = 10.08EE490 pKa = 4.01ARR492 pKa = 11.84LTIICPYY499 pKa = 7.61TQPPLTDD506 pKa = 3.9PEE508 pKa = 4.15NVDD511 pKa = 3.65MGFIPYY517 pKa = 10.25DD518 pKa = 3.64FNFGNGVMPDD528 pKa = 3.78GTPYY532 pKa = 10.4IPIEE536 pKa = 4.54CRR538 pKa = 11.84MKK540 pKa = 9.71WYY542 pKa = 9.45PCMFHH547 pKa = 5.86QQNFMNSIVSCGPFAYY563 pKa = 10.15QGDD566 pKa = 4.25EE567 pKa = 4.02KK568 pKa = 11.37SVVLTAKK575 pKa = 10.53YY576 pKa = 10.33KK577 pKa = 10.82FNFLFGGNPTPEE589 pKa = 3.92QTIKK593 pKa = 10.92DD594 pKa = 3.84PCQQPTFEE602 pKa = 4.81MPGSSGLPRR611 pKa = 11.84DD612 pKa = 4.01VQIEE616 pKa = 4.17DD617 pKa = 3.66PQLLHH622 pKa = 6.3EE623 pKa = 5.35GYY625 pKa = 10.74YY626 pKa = 9.99FRR628 pKa = 11.84AWDD631 pKa = 4.09LRR633 pKa = 11.84RR634 pKa = 11.84GLYY637 pKa = 9.9GEE639 pKa = 4.27KK640 pKa = 10.04AIKK643 pKa = 10.09RR644 pKa = 11.84MRR646 pKa = 11.84EE647 pKa = 3.67QPIPPEE653 pKa = 4.4FFAGPPKK660 pKa = 10.29KK661 pKa = 10.48SRR663 pKa = 11.84FEE665 pKa = 4.08VPANVADD672 pKa = 4.76DD673 pKa = 4.11YY674 pKa = 11.83SSKK677 pKa = 8.29EE678 pKa = 4.31QKK680 pKa = 7.98WHH682 pKa = 6.72PWPDD686 pKa = 3.4SPGAEE691 pKa = 3.81AQSSDD696 pKa = 3.73EE697 pKa = 4.21EE698 pKa = 4.31EE699 pKa = 5.44KK700 pKa = 10.86EE701 pKa = 4.14VQTPILQHH709 pKa = 5.53QLRR712 pKa = 11.84EE713 pKa = 4.01HH714 pKa = 6.2LRR716 pKa = 11.84EE717 pKa = 3.98QKK719 pKa = 10.38VLKK722 pKa = 10.4SQIKK726 pKa = 9.11FVIKK730 pKa = 10.36QLMKK734 pKa = 9.24TQSHH738 pKa = 5.77LHH740 pKa = 4.93VPYY743 pKa = 10.88YY744 pKa = 9.74PP745 pKa = 3.96
MM1 pKa = 7.0WWNRR5 pKa = 11.84WKK7 pKa = 10.57RR8 pKa = 11.84YY9 pKa = 6.18RR10 pKa = 11.84WRR12 pKa = 11.84PLRR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84TYY20 pKa = 9.03RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PLRR27 pKa = 11.84WRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84GAARR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84VRR43 pKa = 11.84RR44 pKa = 11.84WRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AWGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84YY57 pKa = 9.38IRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84LRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.74KK69 pKa = 10.57IPITQWNPSVVRR81 pKa = 11.84KK82 pKa = 9.76CVVTGYY88 pKa = 10.29IPLLICGTGTTGTTYY103 pKa = 11.21KK104 pKa = 10.81NYY106 pKa = 10.12GSHH109 pKa = 6.29AADD112 pKa = 3.47YY113 pKa = 10.8KK114 pKa = 11.06KK115 pKa = 10.68YY116 pKa = 10.53DD117 pKa = 3.87PFGGGYY123 pKa = 7.98STMQFTLEE131 pKa = 4.07TLFDD135 pKa = 3.37EE136 pKa = 4.96WVKK139 pKa = 10.68HH140 pKa = 4.8RR141 pKa = 11.84CTWSRR146 pKa = 11.84SNKK149 pKa = 9.64DD150 pKa = 3.42LEE152 pKa = 4.23LVRR155 pKa = 11.84YY156 pKa = 9.79LGGSFTLYY164 pKa = 10.2RR165 pKa = 11.84HH166 pKa = 6.36PKK168 pKa = 8.45CDD170 pKa = 4.42FIFRR174 pKa = 11.84YY175 pKa = 9.0NRR177 pKa = 11.84KK178 pKa = 8.92PPFKK182 pKa = 9.28DD183 pKa = 3.34TQITGPSLHH192 pKa = 7.02PGILMTRR199 pKa = 11.84RR200 pKa = 11.84KK201 pKa = 9.98KK202 pKa = 10.72KK203 pKa = 9.68IIKK206 pKa = 10.22SFLTRR211 pKa = 11.84PRR213 pKa = 11.84GRR215 pKa = 11.84ATKK218 pKa = 10.09RR219 pKa = 11.84VRR221 pKa = 11.84FKK223 pKa = 11.11PPTLYY228 pKa = 9.54TDD230 pKa = 2.56KK231 pKa = 10.66WYY233 pKa = 9.67FQKK236 pKa = 10.7DD237 pKa = 3.24ICNLPLVNIAASACSLRR254 pKa = 11.84FPFCSPQTDD263 pKa = 3.56NTCVNFQVLSKK274 pKa = 10.62RR275 pKa = 11.84FNTMLSISPDD285 pKa = 3.77YY286 pKa = 10.34PKK288 pKa = 10.91KK289 pKa = 10.8NYY291 pKa = 10.4DD292 pKa = 3.85SFVKK296 pKa = 10.0TYY298 pKa = 10.81LKK300 pKa = 10.43NAQEE304 pKa = 4.19HH305 pKa = 5.36WNQNKK310 pKa = 10.23KK311 pKa = 8.73GTLDD315 pKa = 3.54NYY317 pKa = 10.92NDD319 pKa = 4.12PNRR322 pKa = 11.84LLTVFNTFKK331 pKa = 10.63TEE333 pKa = 3.42EE334 pKa = 4.08HH335 pKa = 6.84LYY337 pKa = 10.89DD338 pKa = 4.33PSAKK342 pKa = 8.26EE343 pKa = 4.21TKK345 pKa = 9.14NTTNTDD351 pKa = 3.27AAGNKK356 pKa = 9.07YY357 pKa = 8.63NTVTSLWGDD366 pKa = 3.6YY367 pKa = 9.88IYY369 pKa = 11.16KK370 pKa = 8.65EE371 pKa = 4.21TVVDD375 pKa = 3.83NFINNAKK382 pKa = 10.47NYY384 pKa = 9.58FEE386 pKa = 4.21SRR388 pKa = 11.84KK389 pKa = 10.5GNIVMSNEE397 pKa = 3.84YY398 pKa = 10.46LNHH401 pKa = 5.55KK402 pKa = 8.56TGLYY406 pKa = 10.5SPIFLSNSRR415 pKa = 11.84LSPDD419 pKa = 3.13FPGFYY424 pKa = 9.38IDD426 pKa = 4.78VLYY429 pKa = 11.32NPANDD434 pKa = 3.59KK435 pKa = 11.5GIGNKK440 pKa = 7.93IWMDD444 pKa = 3.17WCTKK448 pKa = 10.56NDD450 pKa = 3.7STWRR454 pKa = 11.84DD455 pKa = 3.6TPQKK459 pKa = 10.46LPVVDD464 pKa = 3.74IPLWAALLGYY474 pKa = 10.11SDD476 pKa = 4.93YY477 pKa = 9.93CTKK480 pKa = 10.7YY481 pKa = 10.83FNDD484 pKa = 3.38KK485 pKa = 11.13GLIKK489 pKa = 10.08EE490 pKa = 4.01ARR492 pKa = 11.84LTIICPYY499 pKa = 7.61TQPPLTDD506 pKa = 3.9PEE508 pKa = 4.15NVDD511 pKa = 3.65MGFIPYY517 pKa = 10.25DD518 pKa = 3.64FNFGNGVMPDD528 pKa = 3.78GTPYY532 pKa = 10.4IPIEE536 pKa = 4.54CRR538 pKa = 11.84MKK540 pKa = 9.71WYY542 pKa = 9.45PCMFHH547 pKa = 5.86QQNFMNSIVSCGPFAYY563 pKa = 10.15QGDD566 pKa = 4.25EE567 pKa = 4.02KK568 pKa = 11.37SVVLTAKK575 pKa = 10.53YY576 pKa = 10.33KK577 pKa = 10.82FNFLFGGNPTPEE589 pKa = 3.92QTIKK593 pKa = 10.92DD594 pKa = 3.84PCQQPTFEE602 pKa = 4.81MPGSSGLPRR611 pKa = 11.84DD612 pKa = 4.01VQIEE616 pKa = 4.17DD617 pKa = 3.66PQLLHH622 pKa = 6.3EE623 pKa = 5.35GYY625 pKa = 10.74YY626 pKa = 9.99FRR628 pKa = 11.84AWDD631 pKa = 4.09LRR633 pKa = 11.84RR634 pKa = 11.84GLYY637 pKa = 9.9GEE639 pKa = 4.27KK640 pKa = 10.04AIKK643 pKa = 10.09RR644 pKa = 11.84MRR646 pKa = 11.84EE647 pKa = 3.67QPIPPEE653 pKa = 4.4FFAGPPKK660 pKa = 10.29KK661 pKa = 10.48SRR663 pKa = 11.84FEE665 pKa = 4.08VPANVADD672 pKa = 4.76DD673 pKa = 4.11YY674 pKa = 11.83SSKK677 pKa = 8.29EE678 pKa = 4.31QKK680 pKa = 7.98WHH682 pKa = 6.72PWPDD686 pKa = 3.4SPGAEE691 pKa = 3.81AQSSDD696 pKa = 3.73EE697 pKa = 4.21EE698 pKa = 4.31EE699 pKa = 5.44KK700 pKa = 10.86EE701 pKa = 4.14VQTPILQHH709 pKa = 5.53QLRR712 pKa = 11.84EE713 pKa = 4.01HH714 pKa = 6.2LRR716 pKa = 11.84EE717 pKa = 3.98QKK719 pKa = 10.38VLKK722 pKa = 10.4SQIKK726 pKa = 9.11FVIKK730 pKa = 10.36QLMKK734 pKa = 9.24TQSHH738 pKa = 5.77LHH740 pKa = 4.93VPYY743 pKa = 10.88YY744 pKa = 9.74PP745 pKa = 3.96
Molecular weight: 88.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1416 |
125 |
745 |
354.0 |
40.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.638 ± 2.196 | 2.472 ± 0.374 |
5.367 ± 0.321 | 4.661 ± 0.474 |
3.602 ± 1.039 | 7.203 ± 1.517 |
1.624 ± 0.179 | 3.46 ± 0.824 |
5.932 ± 1.46 | 6.356 ± 0.381 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.412 ± 0.321 | 4.167 ± 1.159 |
9.251 ± 1.174 | 4.096 ± 0.357 |
9.463 ± 0.587 | 8.121 ± 2.386 |
5.932 ± 0.696 | 4.096 ± 0.16 |
2.189 ± 0.532 | 3.955 ± 1.12 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
