
Nocardioides currus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
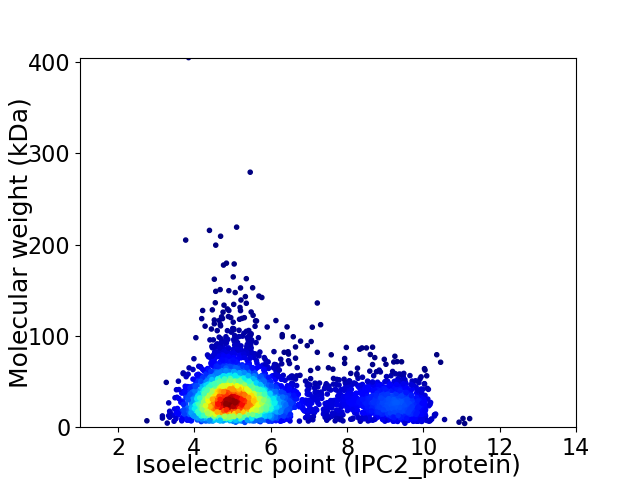
Virtual 2D-PAGE plot for 4242 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R7YXU4|A0A2R7YXU4_9ACTN Uridylate kinase OS=Nocardioides currus OX=2133958 GN=pyrH PE=3 SV=1
MM1 pKa = 7.13TVDD4 pKa = 4.37AVDD7 pKa = 4.17DD8 pKa = 3.72QAVANDD14 pKa = 3.96DD15 pKa = 3.78EE16 pKa = 4.56LTIEE20 pKa = 4.23EE21 pKa = 4.84DD22 pKa = 3.4ADD24 pKa = 3.66ATEE27 pKa = 4.32VDD29 pKa = 4.05VLANDD34 pKa = 3.81TDD36 pKa = 4.15VEE38 pKa = 4.23GDD40 pKa = 4.08PIEE43 pKa = 4.23VTAVGDD49 pKa = 4.03AEE51 pKa = 4.86HH52 pKa = 6.35GTVTLVDD59 pKa = 3.77GVVTYY64 pKa = 9.23TPNANYY70 pKa = 10.65NGTDD74 pKa = 3.07SFTYY78 pKa = 9.81TVTDD82 pKa = 3.89GATATVAVTVNAVDD96 pKa = 4.49DD97 pKa = 4.5KK98 pKa = 11.36PVAHH102 pKa = 7.44DD103 pKa = 4.4DD104 pKa = 3.86EE105 pKa = 4.95VTVTEE110 pKa = 4.99DD111 pKa = 3.26DD112 pKa = 3.65PAAVVDD118 pKa = 4.23VLANDD123 pKa = 3.7TDD125 pKa = 3.8IDD127 pKa = 4.03GGPKK131 pKa = 9.61HH132 pKa = 5.81VASVGAAAHH141 pKa = 5.38GTVTLVDD148 pKa = 3.82GVVSYY153 pKa = 10.98RR154 pKa = 11.84PNANHH159 pKa = 7.02AGSDD163 pKa = 3.53SFTYY167 pKa = 9.11TVNGGSSATVDD178 pKa = 3.23ITVSAVDD185 pKa = 3.85DD186 pKa = 4.51APVAVADD193 pKa = 4.01SAAVAEE199 pKa = 4.82DD200 pKa = 3.85ASATAINVLANDD212 pKa = 3.84TDD214 pKa = 3.81IDD216 pKa = 4.13GGPKK220 pKa = 10.3AIMSATNPAHH230 pKa = 5.18GTVVITGGGSGLTYY244 pKa = 10.45RR245 pKa = 11.84PAANYY250 pKa = 10.19NGADD254 pKa = 3.33SFTYY258 pKa = 9.39TLNGGSTATVSITVTAVDD276 pKa = 4.16DD277 pKa = 4.42APVAVNDD284 pKa = 4.39AYY286 pKa = 9.8TVSEE290 pKa = 4.61DD291 pKa = 3.93SNATTLPVLANDD303 pKa = 4.05ADD305 pKa = 3.98IDD307 pKa = 4.16GGPKK311 pKa = 9.97TITAVTQPANGTVTITGGGTGLTYY335 pKa = 10.66KK336 pKa = 10.56PATNYY341 pKa = 7.41FTPLNTPSTFTYY353 pKa = 9.95TLDD356 pKa = 3.83GGSTATVSMTVSPVADD372 pKa = 3.39NGASNNPTIEE382 pKa = 4.09VGAEE386 pKa = 3.74TTVYY390 pKa = 10.52KK391 pKa = 10.53IGTAPVVIAPNLALTHH407 pKa = 6.75PDD409 pKa = 2.8WPSFSGATVRR419 pKa = 11.84IPGTAQAGDD428 pKa = 3.42VIGFVNQNGIVGSYY442 pKa = 11.34SNGTLTLTGVASPAAYY458 pKa = 9.37QAALRR463 pKa = 11.84TVTFKK468 pKa = 10.58ATSTPEE474 pKa = 3.57TFPIVFTVTDD484 pKa = 4.05GATPARR490 pKa = 11.84SATDD494 pKa = 3.29NRR496 pKa = 11.84SVQITSS502 pKa = 3.02
MM1 pKa = 7.13TVDD4 pKa = 4.37AVDD7 pKa = 4.17DD8 pKa = 3.72QAVANDD14 pKa = 3.96DD15 pKa = 3.78EE16 pKa = 4.56LTIEE20 pKa = 4.23EE21 pKa = 4.84DD22 pKa = 3.4ADD24 pKa = 3.66ATEE27 pKa = 4.32VDD29 pKa = 4.05VLANDD34 pKa = 3.81TDD36 pKa = 4.15VEE38 pKa = 4.23GDD40 pKa = 4.08PIEE43 pKa = 4.23VTAVGDD49 pKa = 4.03AEE51 pKa = 4.86HH52 pKa = 6.35GTVTLVDD59 pKa = 3.77GVVTYY64 pKa = 9.23TPNANYY70 pKa = 10.65NGTDD74 pKa = 3.07SFTYY78 pKa = 9.81TVTDD82 pKa = 3.89GATATVAVTVNAVDD96 pKa = 4.49DD97 pKa = 4.5KK98 pKa = 11.36PVAHH102 pKa = 7.44DD103 pKa = 4.4DD104 pKa = 3.86EE105 pKa = 4.95VTVTEE110 pKa = 4.99DD111 pKa = 3.26DD112 pKa = 3.65PAAVVDD118 pKa = 4.23VLANDD123 pKa = 3.7TDD125 pKa = 3.8IDD127 pKa = 4.03GGPKK131 pKa = 9.61HH132 pKa = 5.81VASVGAAAHH141 pKa = 5.38GTVTLVDD148 pKa = 3.82GVVSYY153 pKa = 10.98RR154 pKa = 11.84PNANHH159 pKa = 7.02AGSDD163 pKa = 3.53SFTYY167 pKa = 9.11TVNGGSSATVDD178 pKa = 3.23ITVSAVDD185 pKa = 3.85DD186 pKa = 4.51APVAVADD193 pKa = 4.01SAAVAEE199 pKa = 4.82DD200 pKa = 3.85ASATAINVLANDD212 pKa = 3.84TDD214 pKa = 3.81IDD216 pKa = 4.13GGPKK220 pKa = 10.3AIMSATNPAHH230 pKa = 5.18GTVVITGGGSGLTYY244 pKa = 10.45RR245 pKa = 11.84PAANYY250 pKa = 10.19NGADD254 pKa = 3.33SFTYY258 pKa = 9.39TLNGGSTATVSITVTAVDD276 pKa = 4.16DD277 pKa = 4.42APVAVNDD284 pKa = 4.39AYY286 pKa = 9.8TVSEE290 pKa = 4.61DD291 pKa = 3.93SNATTLPVLANDD303 pKa = 4.05ADD305 pKa = 3.98IDD307 pKa = 4.16GGPKK311 pKa = 9.97TITAVTQPANGTVTITGGGTGLTYY335 pKa = 10.66KK336 pKa = 10.56PATNYY341 pKa = 7.41FTPLNTPSTFTYY353 pKa = 9.95TLDD356 pKa = 3.83GGSTATVSMTVSPVADD372 pKa = 3.39NGASNNPTIEE382 pKa = 4.09VGAEE386 pKa = 3.74TTVYY390 pKa = 10.52KK391 pKa = 10.53IGTAPVVIAPNLALTHH407 pKa = 6.75PDD409 pKa = 2.8WPSFSGATVRR419 pKa = 11.84IPGTAQAGDD428 pKa = 3.42VIGFVNQNGIVGSYY442 pKa = 11.34SNGTLTLTGVASPAAYY458 pKa = 9.37QAALRR463 pKa = 11.84TVTFKK468 pKa = 10.58ATSTPEE474 pKa = 3.57TFPIVFTVTDD484 pKa = 4.05GATPARR490 pKa = 11.84SATDD494 pKa = 3.29NRR496 pKa = 11.84SVQITSS502 pKa = 3.02
Molecular weight: 50.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R7YT29|A0A2R7YT29_9ACTN Protocatechuate 3 4-dioxygenase subunit alpha OS=Nocardioides currus OX=2133958 GN=pcaG PE=3 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84CPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84STRR21 pKa = 11.84PRR23 pKa = 11.84ARR25 pKa = 11.84PRR27 pKa = 11.84SRR29 pKa = 11.84PTRR32 pKa = 11.84PSRR35 pKa = 11.84ASRR38 pKa = 11.84SPRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84PSPTQPLRR53 pKa = 11.84PPRR56 pKa = 11.84GRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84PTRR63 pKa = 11.84AIPFPEE69 pKa = 3.76TRR71 pKa = 11.84VFLGPRR77 pKa = 3.37
MM1 pKa = 7.64RR2 pKa = 11.84CPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84STRR21 pKa = 11.84PRR23 pKa = 11.84ARR25 pKa = 11.84PRR27 pKa = 11.84SRR29 pKa = 11.84PTRR32 pKa = 11.84PSRR35 pKa = 11.84ASRR38 pKa = 11.84SPRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84PSPTQPLRR53 pKa = 11.84PPRR56 pKa = 11.84GRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84PTRR63 pKa = 11.84AIPFPEE69 pKa = 3.76TRR71 pKa = 11.84VFLGPRR77 pKa = 3.37
Molecular weight: 9.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1384144 |
33 |
4116 |
326.3 |
34.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.943 ± 0.049 | 0.709 ± 0.009 |
6.763 ± 0.036 | 5.652 ± 0.041 |
2.839 ± 0.022 | 9.1 ± 0.033 |
2.206 ± 0.021 | 3.55 ± 0.025 |
1.971 ± 0.03 | 10.126 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.886 ± 0.016 | 1.784 ± 0.025 |
5.509 ± 0.023 | 2.679 ± 0.019 |
7.462 ± 0.039 | 5.521 ± 0.029 |
6.359 ± 0.047 | 9.477 ± 0.038 |
1.574 ± 0.018 | 1.888 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
