
Phenylobacterium zucineum (strain HLK1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Phenylobacterium; Phenylobacterium zucineum
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
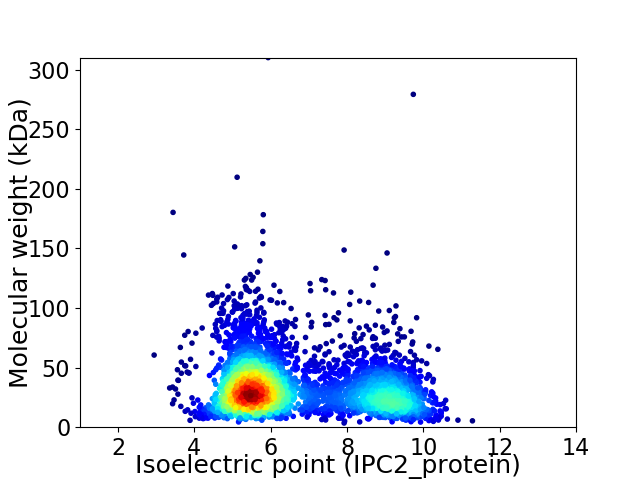
Virtual 2D-PAGE plot for 3838 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
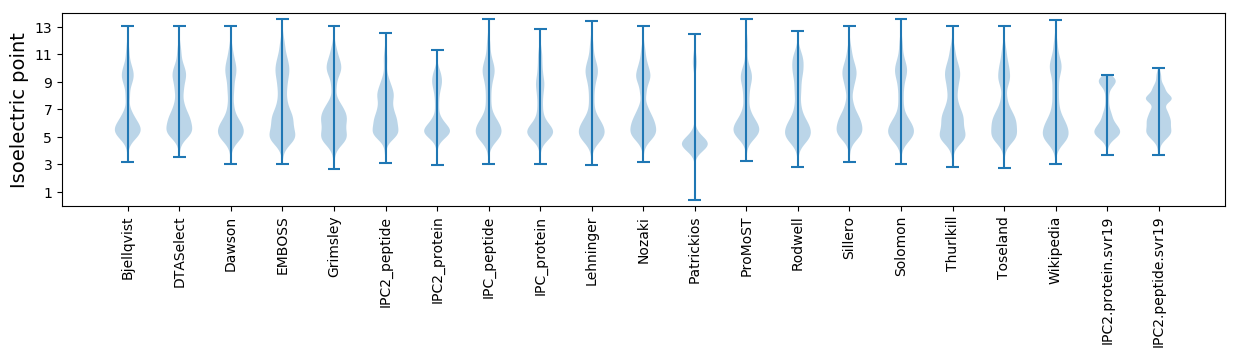
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B4RDL3|B4RDL3_PHEZH Integral membrane protein OS=Phenylobacterium zucineum (strain HLK1) OX=450851 GN=PHZ_c1992 PE=4 SV=1
MM1 pKa = 7.29SVGVSVPQSGDD12 pKa = 3.66PIVDD16 pKa = 3.4GLLFGSKK23 pKa = 8.33WSGTSLTFSFPASVADD39 pKa = 3.9LADD42 pKa = 3.61YY43 pKa = 7.34ATPLDD48 pKa = 3.59PTYY51 pKa = 10.88FGVLEE56 pKa = 4.32AAGQTAVRR64 pKa = 11.84AALARR69 pKa = 11.84WAAVAAVTFTEE80 pKa = 4.54VAPSAAADD88 pKa = 3.57LRR90 pKa = 11.84IYY92 pKa = 10.51HH93 pKa = 6.46YY94 pKa = 11.26DD95 pKa = 3.78DD96 pKa = 3.66GTLFTARR103 pKa = 11.84VVAFPDD109 pKa = 3.46ATPEE113 pKa = 4.31AGDD116 pKa = 3.75LQFGSMFVGPDD127 pKa = 3.28WSPGNYY133 pKa = 9.36HH134 pKa = 6.48YY135 pKa = 9.7FTVVHH140 pKa = 6.83EE141 pKa = 4.6IGHH144 pKa = 6.16ALGLKK149 pKa = 9.78HH150 pKa = 6.8PHH152 pKa = 6.38DD153 pKa = 4.42TPNGFPAADD162 pKa = 3.58SAVDD166 pKa = 3.93AVAEE170 pKa = 4.51TIMSYY175 pKa = 10.42RR176 pKa = 11.84SYY178 pKa = 11.33VGGPMGGYY186 pKa = 9.49SLQAGSYY193 pKa = 8.98PVGPMPSDD201 pKa = 3.11IAAIQHH207 pKa = 6.77LYY209 pKa = 9.01GPNWATNAGDD219 pKa = 3.44TTYY222 pKa = 10.8TFDD225 pKa = 4.01PSASVIFQAVWDD237 pKa = 4.14GGGTDD242 pKa = 3.63TYY244 pKa = 11.78NFASYY249 pKa = 10.59AADD252 pKa = 3.35LHH254 pKa = 6.5IDD256 pKa = 3.73LRR258 pKa = 11.84AGEE261 pKa = 4.07WSDD264 pKa = 5.33LGGQYY269 pKa = 11.15AVLDD273 pKa = 3.88TGDD276 pKa = 3.2ASIRR280 pKa = 11.84PSGNVANPHH289 pKa = 7.02LYY291 pKa = 10.38QGDD294 pKa = 3.91LRR296 pKa = 11.84SLIEE300 pKa = 3.94NAVGGSGDD308 pKa = 3.72DD309 pKa = 3.46TLTGNQAANTLTGGAGDD326 pKa = 4.37DD327 pKa = 4.06SLVGGDD333 pKa = 4.2GADD336 pKa = 3.46TLVGGSGNDD345 pKa = 3.66TLSGGAGSDD354 pKa = 3.15RR355 pKa = 11.84FTLGGGGDD363 pKa = 3.96VIADD367 pKa = 4.03FLAGDD372 pKa = 4.41RR373 pKa = 11.84IVAQGAVAGDD383 pKa = 3.6LTFDD387 pKa = 3.45LTGEE391 pKa = 4.28ILTIDD396 pKa = 3.91PDD398 pKa = 4.16GAGAAASFTVTVTGGLLSGYY418 pKa = 8.83EE419 pKa = 4.14LVAEE423 pKa = 4.42ADD425 pKa = 3.99GGLVWKK431 pKa = 10.08ALPPPPEE438 pKa = 4.61PPPPGPTHH446 pKa = 6.51TDD448 pKa = 3.16QPGEE452 pKa = 3.95GSDD455 pKa = 3.94FVVLSQEE462 pKa = 3.89GTTFAGAGADD472 pKa = 3.19WVLGSIEE479 pKa = 4.17GDD481 pKa = 2.97WVHH484 pKa = 6.7GNQGDD489 pKa = 4.26DD490 pKa = 3.5YY491 pKa = 10.89LHH493 pKa = 7.22GYY495 pKa = 8.43GAADD499 pKa = 3.36TVYY502 pKa = 10.56GGQGDD507 pKa = 3.85DD508 pKa = 3.98RR509 pKa = 11.84VFGDD513 pKa = 3.93EE514 pKa = 4.83GDD516 pKa = 3.73DD517 pKa = 4.76AIYY520 pKa = 11.06GDD522 pKa = 4.25MGSDD526 pKa = 3.24RR527 pKa = 11.84LDD529 pKa = 3.55GGTGDD534 pKa = 4.91DD535 pKa = 4.15VAHH538 pKa = 7.07GGPGDD543 pKa = 3.84DD544 pKa = 4.01VVEE547 pKa = 4.66GGQGADD553 pKa = 3.51LLYY556 pKa = 10.76GGKK559 pKa = 9.96DD560 pKa = 3.11ADD562 pKa = 4.34TVSGGAGDD570 pKa = 3.59DD571 pKa = 4.36WIWGDD576 pKa = 4.48LGDD579 pKa = 5.26DD580 pKa = 3.96RR581 pKa = 11.84LSGGAGADD589 pKa = 2.78RR590 pKa = 11.84FAFVAGYY597 pKa = 10.48AGNDD601 pKa = 3.44VIADD605 pKa = 4.28FSAADD610 pKa = 3.49GDD612 pKa = 4.24RR613 pKa = 11.84IEE615 pKa = 4.37LRR617 pKa = 11.84GVEE620 pKa = 4.69DD621 pKa = 4.85FAALTIGSSAAGATILLPTGEE642 pKa = 4.85VITLLGVPGASLGASDD658 pKa = 5.27FLFGG662 pKa = 5.0
MM1 pKa = 7.29SVGVSVPQSGDD12 pKa = 3.66PIVDD16 pKa = 3.4GLLFGSKK23 pKa = 8.33WSGTSLTFSFPASVADD39 pKa = 3.9LADD42 pKa = 3.61YY43 pKa = 7.34ATPLDD48 pKa = 3.59PTYY51 pKa = 10.88FGVLEE56 pKa = 4.32AAGQTAVRR64 pKa = 11.84AALARR69 pKa = 11.84WAAVAAVTFTEE80 pKa = 4.54VAPSAAADD88 pKa = 3.57LRR90 pKa = 11.84IYY92 pKa = 10.51HH93 pKa = 6.46YY94 pKa = 11.26DD95 pKa = 3.78DD96 pKa = 3.66GTLFTARR103 pKa = 11.84VVAFPDD109 pKa = 3.46ATPEE113 pKa = 4.31AGDD116 pKa = 3.75LQFGSMFVGPDD127 pKa = 3.28WSPGNYY133 pKa = 9.36HH134 pKa = 6.48YY135 pKa = 9.7FTVVHH140 pKa = 6.83EE141 pKa = 4.6IGHH144 pKa = 6.16ALGLKK149 pKa = 9.78HH150 pKa = 6.8PHH152 pKa = 6.38DD153 pKa = 4.42TPNGFPAADD162 pKa = 3.58SAVDD166 pKa = 3.93AVAEE170 pKa = 4.51TIMSYY175 pKa = 10.42RR176 pKa = 11.84SYY178 pKa = 11.33VGGPMGGYY186 pKa = 9.49SLQAGSYY193 pKa = 8.98PVGPMPSDD201 pKa = 3.11IAAIQHH207 pKa = 6.77LYY209 pKa = 9.01GPNWATNAGDD219 pKa = 3.44TTYY222 pKa = 10.8TFDD225 pKa = 4.01PSASVIFQAVWDD237 pKa = 4.14GGGTDD242 pKa = 3.63TYY244 pKa = 11.78NFASYY249 pKa = 10.59AADD252 pKa = 3.35LHH254 pKa = 6.5IDD256 pKa = 3.73LRR258 pKa = 11.84AGEE261 pKa = 4.07WSDD264 pKa = 5.33LGGQYY269 pKa = 11.15AVLDD273 pKa = 3.88TGDD276 pKa = 3.2ASIRR280 pKa = 11.84PSGNVANPHH289 pKa = 7.02LYY291 pKa = 10.38QGDD294 pKa = 3.91LRR296 pKa = 11.84SLIEE300 pKa = 3.94NAVGGSGDD308 pKa = 3.72DD309 pKa = 3.46TLTGNQAANTLTGGAGDD326 pKa = 4.37DD327 pKa = 4.06SLVGGDD333 pKa = 4.2GADD336 pKa = 3.46TLVGGSGNDD345 pKa = 3.66TLSGGAGSDD354 pKa = 3.15RR355 pKa = 11.84FTLGGGGDD363 pKa = 3.96VIADD367 pKa = 4.03FLAGDD372 pKa = 4.41RR373 pKa = 11.84IVAQGAVAGDD383 pKa = 3.6LTFDD387 pKa = 3.45LTGEE391 pKa = 4.28ILTIDD396 pKa = 3.91PDD398 pKa = 4.16GAGAAASFTVTVTGGLLSGYY418 pKa = 8.83EE419 pKa = 4.14LVAEE423 pKa = 4.42ADD425 pKa = 3.99GGLVWKK431 pKa = 10.08ALPPPPEE438 pKa = 4.61PPPPGPTHH446 pKa = 6.51TDD448 pKa = 3.16QPGEE452 pKa = 3.95GSDD455 pKa = 3.94FVVLSQEE462 pKa = 3.89GTTFAGAGADD472 pKa = 3.19WVLGSIEE479 pKa = 4.17GDD481 pKa = 2.97WVHH484 pKa = 6.7GNQGDD489 pKa = 4.26DD490 pKa = 3.5YY491 pKa = 10.89LHH493 pKa = 7.22GYY495 pKa = 8.43GAADD499 pKa = 3.36TVYY502 pKa = 10.56GGQGDD507 pKa = 3.85DD508 pKa = 3.98RR509 pKa = 11.84VFGDD513 pKa = 3.93EE514 pKa = 4.83GDD516 pKa = 3.73DD517 pKa = 4.76AIYY520 pKa = 11.06GDD522 pKa = 4.25MGSDD526 pKa = 3.24RR527 pKa = 11.84LDD529 pKa = 3.55GGTGDD534 pKa = 4.91DD535 pKa = 4.15VAHH538 pKa = 7.07GGPGDD543 pKa = 3.84DD544 pKa = 4.01VVEE547 pKa = 4.66GGQGADD553 pKa = 3.51LLYY556 pKa = 10.76GGKK559 pKa = 9.96DD560 pKa = 3.11ADD562 pKa = 4.34TVSGGAGDD570 pKa = 3.59DD571 pKa = 4.36WIWGDD576 pKa = 4.48LGDD579 pKa = 5.26DD580 pKa = 3.96RR581 pKa = 11.84LSGGAGADD589 pKa = 2.78RR590 pKa = 11.84FAFVAGYY597 pKa = 10.48AGNDD601 pKa = 3.44VIADD605 pKa = 4.28FSAADD610 pKa = 3.49GDD612 pKa = 4.24RR613 pKa = 11.84IEE615 pKa = 4.37LRR617 pKa = 11.84GVEE620 pKa = 4.69DD621 pKa = 4.85FAALTIGSSAAGATILLPTGEE642 pKa = 4.85VITLLGVPGASLGASDD658 pKa = 5.27FLFGG662 pKa = 5.0
Molecular weight: 66.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|B4RDX2|PURA_PHEZH Adenylosuccinate synthetase OS=Phenylobacterium zucineum (strain HLK1) OX=450851 GN=purA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.11RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.53GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.02NGQKK29 pKa = 9.43ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.11RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.53GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.02NGQKK29 pKa = 9.43ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1282678 |
32 |
2838 |
334.2 |
35.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.46 ± 0.072 | 0.757 ± 0.011 |
5.577 ± 0.034 | 6.065 ± 0.039 |
3.481 ± 0.024 | 9.251 ± 0.039 |
1.813 ± 0.022 | 3.913 ± 0.027 |
2.727 ± 0.033 | 10.078 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.11 ± 0.018 | 2.061 ± 0.027 |
5.852 ± 0.03 | 3.07 ± 0.02 |
8.168 ± 0.044 | 4.555 ± 0.027 |
4.794 ± 0.031 | 7.731 ± 0.036 |
1.433 ± 0.018 | 2.103 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
