
Middle East respiratory syndrome-related coronavirus (MERS-CoV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Merbecovirus
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
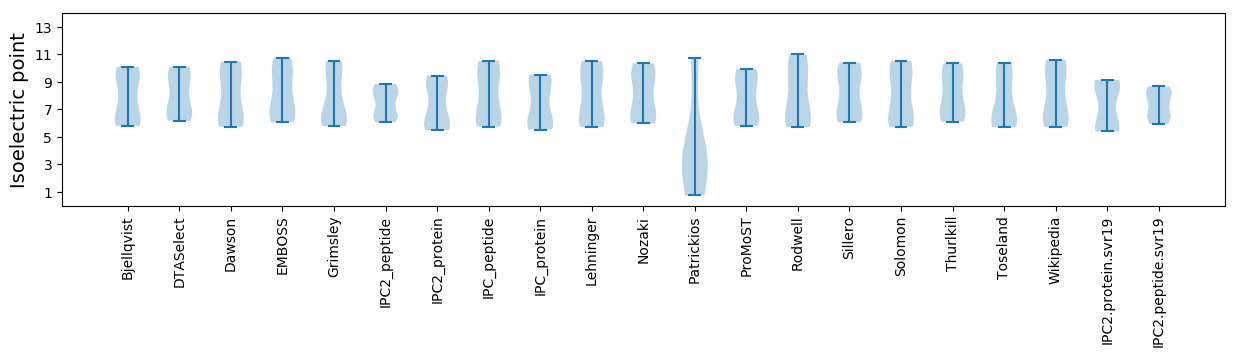
* You can choose from 21 different methods for calculating isoelectric point
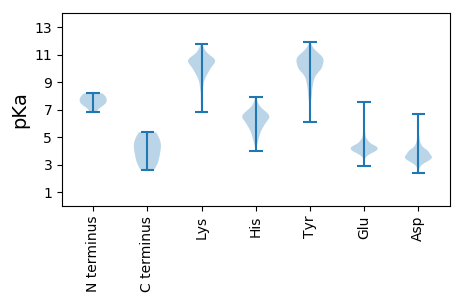
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9UQM6|R9UQM6_MERS NS3D protein OS=Middle East respiratory syndrome-related coronavirus OX=1335626 GN=orf5 PE=4 SV=1
MM1 pKa = 6.86IHH3 pKa = 6.87SVFLLMFLLTPTEE16 pKa = 4.37SYY18 pKa = 11.55VDD20 pKa = 3.55VGPDD24 pKa = 3.43SVKK27 pKa = 10.47SACIEE32 pKa = 3.72VDD34 pKa = 2.94IQQTFFDD41 pKa = 4.38KK42 pKa = 10.29TWPRR46 pKa = 11.84PIDD49 pKa = 3.46VSKK52 pKa = 11.24ADD54 pKa = 4.31GIIYY58 pKa = 8.52PQGRR62 pKa = 11.84TYY64 pKa = 11.55SNITITYY71 pKa = 8.99QGLFPYY77 pKa = 10.39QGDD80 pKa = 3.55HH81 pKa = 6.45GDD83 pKa = 3.5MYY85 pKa = 11.3VYY87 pKa = 10.57SAGHH91 pKa = 5.77ATGTTPQKK99 pKa = 10.83LFVANYY105 pKa = 9.11SQDD108 pKa = 3.09VKK110 pKa = 11.08QFANGFVVRR119 pKa = 11.84IGAAANSTGTVIISPSTSATIRR141 pKa = 11.84KK142 pKa = 9.31IYY144 pKa = 8.72PAFMLGSSVGNFSDD158 pKa = 3.45GKK160 pKa = 8.72MGRR163 pKa = 11.84FFNHH167 pKa = 6.01TLVLLPDD174 pKa = 3.85GCGTLLRR181 pKa = 11.84AFYY184 pKa = 10.55CILEE188 pKa = 4.27PRR190 pKa = 11.84SGNHH194 pKa = 5.77CPAGNSYY201 pKa = 9.97TSFATYY207 pKa = 7.63HH208 pKa = 6.01TPATDD213 pKa = 4.12CSDD216 pKa = 3.09GNYY219 pKa = 9.81NRR221 pKa = 11.84NASLNSFKK229 pKa = 10.94EE230 pKa = 3.91YY231 pKa = 10.79FNLRR235 pKa = 11.84NCTFMYY241 pKa = 9.51TYY243 pKa = 10.63NITEE247 pKa = 4.15DD248 pKa = 4.93EE249 pKa = 3.99ILEE252 pKa = 4.35WFGITQTAQGVHH264 pKa = 6.39LFSSRR269 pKa = 11.84YY270 pKa = 7.82VDD272 pKa = 4.34LYY274 pKa = 9.95GGNMFQFATLPVYY287 pKa = 9.53DD288 pKa = 4.09TIKK291 pKa = 9.6YY292 pKa = 9.93YY293 pKa = 10.88SIIPHH298 pKa = 6.57SIRR301 pKa = 11.84SIQSDD306 pKa = 3.44RR307 pKa = 11.84KK308 pKa = 8.9AWAAFYY314 pKa = 10.51VYY316 pKa = 10.62KK317 pKa = 10.57LQPLTFLLDD326 pKa = 3.94FSVDD330 pKa = 3.3GYY332 pKa = 10.29IRR334 pKa = 11.84RR335 pKa = 11.84AIDD338 pKa = 3.32CGFNDD343 pKa = 5.2LSQLHH348 pKa = 6.59CSYY351 pKa = 11.2EE352 pKa = 4.41SFDD355 pKa = 3.95VEE357 pKa = 4.4SGVYY361 pKa = 9.77SVSSFEE367 pKa = 4.52AKK369 pKa = 10.07PSGSVVEE376 pKa = 3.97QAEE379 pKa = 4.32GVEE382 pKa = 4.81CDD384 pKa = 4.79FSPLLSGTPPQVYY397 pKa = 9.55NFKK400 pKa = 10.72RR401 pKa = 11.84LVFTNCNYY409 pKa = 10.82NLTKK413 pKa = 10.6LLSLFSVNDD422 pKa = 3.83FTCSQISPAAIASNCYY438 pKa = 10.04SSLILDD444 pKa = 3.82YY445 pKa = 10.84FSYY448 pKa = 9.64PLSMKK453 pKa = 10.31SDD455 pKa = 3.95LSVSSAGPISQFNYY469 pKa = 9.2KK470 pKa = 10.04QSFSNPTCLILATVPHH486 pKa = 6.9NLTTITKK493 pKa = 8.1PLKK496 pKa = 9.3YY497 pKa = 10.39SYY499 pKa = 10.58INKK502 pKa = 9.66CSRR505 pKa = 11.84LLSDD509 pKa = 5.19DD510 pKa = 3.58RR511 pKa = 11.84TEE513 pKa = 4.39VPQLVNANQYY523 pKa = 9.07SPCVSIVPSTVWEE536 pKa = 4.6DD537 pKa = 2.68GDD539 pKa = 4.03YY540 pKa = 10.91YY541 pKa = 10.98RR542 pKa = 11.84KK543 pKa = 9.48QLSPLEE549 pKa = 4.3GGGWLVASGSTVAMTEE565 pKa = 3.8QLQMGFGITVQYY577 pKa = 9.09GTDD580 pKa = 3.57TNSVCPKK587 pKa = 10.64LEE589 pKa = 3.84FANDD593 pKa = 3.52TKK595 pKa = 10.75IASQLGNCVEE605 pKa = 3.96YY606 pKa = 10.83SLYY609 pKa = 10.47GVSGRR614 pKa = 11.84GVFQNCTAVGVRR626 pKa = 11.84QQRR629 pKa = 11.84FVYY632 pKa = 10.14DD633 pKa = 4.07AYY635 pKa = 11.32QNLVGYY641 pKa = 10.55YY642 pKa = 9.85SDD644 pKa = 4.75DD645 pKa = 3.3GNYY648 pKa = 9.21YY649 pKa = 10.15CLRR652 pKa = 11.84ACVSVPVSVIYY663 pKa = 10.62DD664 pKa = 3.71KK665 pKa = 10.22EE666 pKa = 4.71TKK668 pKa = 8.48THH670 pKa = 5.81ATLFGSVACEE680 pKa = 4.33HH681 pKa = 6.93ISSTMSQYY689 pKa = 11.08SRR691 pKa = 11.84STRR694 pKa = 11.84SMLKK698 pKa = 10.21RR699 pKa = 11.84RR700 pKa = 11.84DD701 pKa = 3.4STYY704 pKa = 11.16GPLQTPVGCVLGLVNSSLFVEE725 pKa = 4.86DD726 pKa = 4.29CKK728 pKa = 11.38LPLGQSLCALPDD740 pKa = 3.78TPSTLTPRR748 pKa = 11.84SVRR751 pKa = 11.84SVPGEE756 pKa = 3.69MRR758 pKa = 11.84LASIAFNHH766 pKa = 7.19PIQVDD771 pKa = 3.76QLNSSYY777 pKa = 10.78FKK779 pKa = 11.13LSIPTNFSFGVTQEE793 pKa = 4.35YY794 pKa = 10.14IQTTIQKK801 pKa = 8.19VTVDD805 pKa = 3.83CKK807 pKa = 10.97QYY809 pKa = 10.71VCNGFQKK816 pKa = 10.67CEE818 pKa = 3.81QLLRR822 pKa = 11.84EE823 pKa = 4.09YY824 pKa = 11.27GQFCSKK830 pKa = 10.24INQALHH836 pKa = 5.92GANLRR841 pKa = 11.84QDD843 pKa = 3.46DD844 pKa = 4.08SVRR847 pKa = 11.84NLFASVKK854 pKa = 9.66SSQSSPIIPGFGGDD868 pKa = 3.83FNLTLLEE875 pKa = 4.3PVSISTGSRR884 pKa = 11.84SARR887 pKa = 11.84SAIEE891 pKa = 4.69DD892 pKa = 3.78LLFDD896 pKa = 4.46KK897 pKa = 10.28VTIADD902 pKa = 3.68PGYY905 pKa = 8.03MQGYY909 pKa = 9.01DD910 pKa = 3.52DD911 pKa = 6.26CMQQGPASARR921 pKa = 11.84DD922 pKa = 4.26LICAQYY928 pKa = 9.2VAGYY932 pKa = 9.81KK933 pKa = 9.75VLPPLMDD940 pKa = 3.55VNMEE944 pKa = 4.0AAYY947 pKa = 9.29TSSLLGSIAGVGWTAGLSSFAAIPFAQSIFYY978 pKa = 10.26RR979 pKa = 11.84LNGVGITQQVLSEE992 pKa = 4.06NQKK995 pKa = 10.94LIANKK1000 pKa = 9.66FNQALGAMQTGFTTTNEE1017 pKa = 4.13AFRR1020 pKa = 11.84KK1021 pKa = 9.46VQDD1024 pKa = 3.57AVNNNAQALSKK1035 pKa = 10.55LASEE1039 pKa = 5.11LSNTFGAISASIGDD1053 pKa = 4.24IIQRR1057 pKa = 11.84LDD1059 pKa = 3.4VLEE1062 pKa = 4.26QDD1064 pKa = 3.65AQIDD1068 pKa = 3.67RR1069 pKa = 11.84LINGRR1074 pKa = 11.84LTTLNAFVAQQLVRR1088 pKa = 11.84SEE1090 pKa = 4.28SAALSAQLAKK1100 pKa = 10.93DD1101 pKa = 3.6KK1102 pKa = 11.5VNEE1105 pKa = 4.19CVKK1108 pKa = 10.57AQSKK1112 pKa = 10.61RR1113 pKa = 11.84SGFCGQGTHH1122 pKa = 5.83IVSFVVNAPNGLYY1135 pKa = 10.31FMHH1138 pKa = 6.54VGYY1141 pKa = 10.68YY1142 pKa = 8.92PSNHH1146 pKa = 6.15IEE1148 pKa = 4.11VVSAYY1153 pKa = 9.92GLCDD1157 pKa = 3.03AANPTNCIAPVNGYY1171 pKa = 9.08FIKK1174 pKa = 10.17TNNTRR1179 pKa = 11.84IVDD1182 pKa = 3.46EE1183 pKa = 4.15WSYY1186 pKa = 10.86TGSSFYY1192 pKa = 11.03APEE1195 pKa = 5.67PITSLNTKK1203 pKa = 9.23YY1204 pKa = 10.36VAPHH1208 pKa = 4.7VTYY1211 pKa = 11.0QNISTNLPPPLLGNSTGIDD1230 pKa = 3.71FQDD1233 pKa = 3.67EE1234 pKa = 3.86LDD1236 pKa = 3.71EE1237 pKa = 4.43FFKK1240 pKa = 11.2NVSTSIPNFGSLTQINTTLLDD1261 pKa = 3.48LTYY1264 pKa = 11.2EE1265 pKa = 4.16MLSLQQVVKK1274 pKa = 10.52ALNEE1278 pKa = 4.09SYY1280 pKa = 10.82IDD1282 pKa = 3.9LKK1284 pKa = 11.28EE1285 pKa = 4.19LGNYY1289 pKa = 7.68TYY1291 pKa = 11.04YY1292 pKa = 11.18NKK1294 pKa = 9.72WPWYY1298 pKa = 8.04IWLGFIAGLVALALCVFFILCCTGCGTNCMGKK1330 pKa = 9.91LKK1332 pKa = 10.64CNRR1335 pKa = 11.84CCDD1338 pKa = 3.3RR1339 pKa = 11.84YY1340 pKa = 10.78EE1341 pKa = 4.28EE1342 pKa = 4.54YY1343 pKa = 10.79DD1344 pKa = 3.78LEE1346 pKa = 4.13PHH1348 pKa = 6.39KK1349 pKa = 11.21VHH1351 pKa = 6.25VHH1353 pKa = 5.33
MM1 pKa = 6.86IHH3 pKa = 6.87SVFLLMFLLTPTEE16 pKa = 4.37SYY18 pKa = 11.55VDD20 pKa = 3.55VGPDD24 pKa = 3.43SVKK27 pKa = 10.47SACIEE32 pKa = 3.72VDD34 pKa = 2.94IQQTFFDD41 pKa = 4.38KK42 pKa = 10.29TWPRR46 pKa = 11.84PIDD49 pKa = 3.46VSKK52 pKa = 11.24ADD54 pKa = 4.31GIIYY58 pKa = 8.52PQGRR62 pKa = 11.84TYY64 pKa = 11.55SNITITYY71 pKa = 8.99QGLFPYY77 pKa = 10.39QGDD80 pKa = 3.55HH81 pKa = 6.45GDD83 pKa = 3.5MYY85 pKa = 11.3VYY87 pKa = 10.57SAGHH91 pKa = 5.77ATGTTPQKK99 pKa = 10.83LFVANYY105 pKa = 9.11SQDD108 pKa = 3.09VKK110 pKa = 11.08QFANGFVVRR119 pKa = 11.84IGAAANSTGTVIISPSTSATIRR141 pKa = 11.84KK142 pKa = 9.31IYY144 pKa = 8.72PAFMLGSSVGNFSDD158 pKa = 3.45GKK160 pKa = 8.72MGRR163 pKa = 11.84FFNHH167 pKa = 6.01TLVLLPDD174 pKa = 3.85GCGTLLRR181 pKa = 11.84AFYY184 pKa = 10.55CILEE188 pKa = 4.27PRR190 pKa = 11.84SGNHH194 pKa = 5.77CPAGNSYY201 pKa = 9.97TSFATYY207 pKa = 7.63HH208 pKa = 6.01TPATDD213 pKa = 4.12CSDD216 pKa = 3.09GNYY219 pKa = 9.81NRR221 pKa = 11.84NASLNSFKK229 pKa = 10.94EE230 pKa = 3.91YY231 pKa = 10.79FNLRR235 pKa = 11.84NCTFMYY241 pKa = 9.51TYY243 pKa = 10.63NITEE247 pKa = 4.15DD248 pKa = 4.93EE249 pKa = 3.99ILEE252 pKa = 4.35WFGITQTAQGVHH264 pKa = 6.39LFSSRR269 pKa = 11.84YY270 pKa = 7.82VDD272 pKa = 4.34LYY274 pKa = 9.95GGNMFQFATLPVYY287 pKa = 9.53DD288 pKa = 4.09TIKK291 pKa = 9.6YY292 pKa = 9.93YY293 pKa = 10.88SIIPHH298 pKa = 6.57SIRR301 pKa = 11.84SIQSDD306 pKa = 3.44RR307 pKa = 11.84KK308 pKa = 8.9AWAAFYY314 pKa = 10.51VYY316 pKa = 10.62KK317 pKa = 10.57LQPLTFLLDD326 pKa = 3.94FSVDD330 pKa = 3.3GYY332 pKa = 10.29IRR334 pKa = 11.84RR335 pKa = 11.84AIDD338 pKa = 3.32CGFNDD343 pKa = 5.2LSQLHH348 pKa = 6.59CSYY351 pKa = 11.2EE352 pKa = 4.41SFDD355 pKa = 3.95VEE357 pKa = 4.4SGVYY361 pKa = 9.77SVSSFEE367 pKa = 4.52AKK369 pKa = 10.07PSGSVVEE376 pKa = 3.97QAEE379 pKa = 4.32GVEE382 pKa = 4.81CDD384 pKa = 4.79FSPLLSGTPPQVYY397 pKa = 9.55NFKK400 pKa = 10.72RR401 pKa = 11.84LVFTNCNYY409 pKa = 10.82NLTKK413 pKa = 10.6LLSLFSVNDD422 pKa = 3.83FTCSQISPAAIASNCYY438 pKa = 10.04SSLILDD444 pKa = 3.82YY445 pKa = 10.84FSYY448 pKa = 9.64PLSMKK453 pKa = 10.31SDD455 pKa = 3.95LSVSSAGPISQFNYY469 pKa = 9.2KK470 pKa = 10.04QSFSNPTCLILATVPHH486 pKa = 6.9NLTTITKK493 pKa = 8.1PLKK496 pKa = 9.3YY497 pKa = 10.39SYY499 pKa = 10.58INKK502 pKa = 9.66CSRR505 pKa = 11.84LLSDD509 pKa = 5.19DD510 pKa = 3.58RR511 pKa = 11.84TEE513 pKa = 4.39VPQLVNANQYY523 pKa = 9.07SPCVSIVPSTVWEE536 pKa = 4.6DD537 pKa = 2.68GDD539 pKa = 4.03YY540 pKa = 10.91YY541 pKa = 10.98RR542 pKa = 11.84KK543 pKa = 9.48QLSPLEE549 pKa = 4.3GGGWLVASGSTVAMTEE565 pKa = 3.8QLQMGFGITVQYY577 pKa = 9.09GTDD580 pKa = 3.57TNSVCPKK587 pKa = 10.64LEE589 pKa = 3.84FANDD593 pKa = 3.52TKK595 pKa = 10.75IASQLGNCVEE605 pKa = 3.96YY606 pKa = 10.83SLYY609 pKa = 10.47GVSGRR614 pKa = 11.84GVFQNCTAVGVRR626 pKa = 11.84QQRR629 pKa = 11.84FVYY632 pKa = 10.14DD633 pKa = 4.07AYY635 pKa = 11.32QNLVGYY641 pKa = 10.55YY642 pKa = 9.85SDD644 pKa = 4.75DD645 pKa = 3.3GNYY648 pKa = 9.21YY649 pKa = 10.15CLRR652 pKa = 11.84ACVSVPVSVIYY663 pKa = 10.62DD664 pKa = 3.71KK665 pKa = 10.22EE666 pKa = 4.71TKK668 pKa = 8.48THH670 pKa = 5.81ATLFGSVACEE680 pKa = 4.33HH681 pKa = 6.93ISSTMSQYY689 pKa = 11.08SRR691 pKa = 11.84STRR694 pKa = 11.84SMLKK698 pKa = 10.21RR699 pKa = 11.84RR700 pKa = 11.84DD701 pKa = 3.4STYY704 pKa = 11.16GPLQTPVGCVLGLVNSSLFVEE725 pKa = 4.86DD726 pKa = 4.29CKK728 pKa = 11.38LPLGQSLCALPDD740 pKa = 3.78TPSTLTPRR748 pKa = 11.84SVRR751 pKa = 11.84SVPGEE756 pKa = 3.69MRR758 pKa = 11.84LASIAFNHH766 pKa = 7.19PIQVDD771 pKa = 3.76QLNSSYY777 pKa = 10.78FKK779 pKa = 11.13LSIPTNFSFGVTQEE793 pKa = 4.35YY794 pKa = 10.14IQTTIQKK801 pKa = 8.19VTVDD805 pKa = 3.83CKK807 pKa = 10.97QYY809 pKa = 10.71VCNGFQKK816 pKa = 10.67CEE818 pKa = 3.81QLLRR822 pKa = 11.84EE823 pKa = 4.09YY824 pKa = 11.27GQFCSKK830 pKa = 10.24INQALHH836 pKa = 5.92GANLRR841 pKa = 11.84QDD843 pKa = 3.46DD844 pKa = 4.08SVRR847 pKa = 11.84NLFASVKK854 pKa = 9.66SSQSSPIIPGFGGDD868 pKa = 3.83FNLTLLEE875 pKa = 4.3PVSISTGSRR884 pKa = 11.84SARR887 pKa = 11.84SAIEE891 pKa = 4.69DD892 pKa = 3.78LLFDD896 pKa = 4.46KK897 pKa = 10.28VTIADD902 pKa = 3.68PGYY905 pKa = 8.03MQGYY909 pKa = 9.01DD910 pKa = 3.52DD911 pKa = 6.26CMQQGPASARR921 pKa = 11.84DD922 pKa = 4.26LICAQYY928 pKa = 9.2VAGYY932 pKa = 9.81KK933 pKa = 9.75VLPPLMDD940 pKa = 3.55VNMEE944 pKa = 4.0AAYY947 pKa = 9.29TSSLLGSIAGVGWTAGLSSFAAIPFAQSIFYY978 pKa = 10.26RR979 pKa = 11.84LNGVGITQQVLSEE992 pKa = 4.06NQKK995 pKa = 10.94LIANKK1000 pKa = 9.66FNQALGAMQTGFTTTNEE1017 pKa = 4.13AFRR1020 pKa = 11.84KK1021 pKa = 9.46VQDD1024 pKa = 3.57AVNNNAQALSKK1035 pKa = 10.55LASEE1039 pKa = 5.11LSNTFGAISASIGDD1053 pKa = 4.24IIQRR1057 pKa = 11.84LDD1059 pKa = 3.4VLEE1062 pKa = 4.26QDD1064 pKa = 3.65AQIDD1068 pKa = 3.67RR1069 pKa = 11.84LINGRR1074 pKa = 11.84LTTLNAFVAQQLVRR1088 pKa = 11.84SEE1090 pKa = 4.28SAALSAQLAKK1100 pKa = 10.93DD1101 pKa = 3.6KK1102 pKa = 11.5VNEE1105 pKa = 4.19CVKK1108 pKa = 10.57AQSKK1112 pKa = 10.61RR1113 pKa = 11.84SGFCGQGTHH1122 pKa = 5.83IVSFVVNAPNGLYY1135 pKa = 10.31FMHH1138 pKa = 6.54VGYY1141 pKa = 10.68YY1142 pKa = 8.92PSNHH1146 pKa = 6.15IEE1148 pKa = 4.11VVSAYY1153 pKa = 9.92GLCDD1157 pKa = 3.03AANPTNCIAPVNGYY1171 pKa = 9.08FIKK1174 pKa = 10.17TNNTRR1179 pKa = 11.84IVDD1182 pKa = 3.46EE1183 pKa = 4.15WSYY1186 pKa = 10.86TGSSFYY1192 pKa = 11.03APEE1195 pKa = 5.67PITSLNTKK1203 pKa = 9.23YY1204 pKa = 10.36VAPHH1208 pKa = 4.7VTYY1211 pKa = 11.0QNISTNLPPPLLGNSTGIDD1230 pKa = 3.71FQDD1233 pKa = 3.67EE1234 pKa = 3.86LDD1236 pKa = 3.71EE1237 pKa = 4.43FFKK1240 pKa = 11.2NVSTSIPNFGSLTQINTTLLDD1261 pKa = 3.48LTYY1264 pKa = 11.2EE1265 pKa = 4.16MLSLQQVVKK1274 pKa = 10.52ALNEE1278 pKa = 4.09SYY1280 pKa = 10.82IDD1282 pKa = 3.9LKK1284 pKa = 11.28EE1285 pKa = 4.19LGNYY1289 pKa = 7.68TYY1291 pKa = 11.04YY1292 pKa = 11.18NKK1294 pKa = 9.72WPWYY1298 pKa = 8.04IWLGFIAGLVALALCVFFILCCTGCGTNCMGKK1330 pKa = 9.91LKK1332 pKa = 10.64CNRR1335 pKa = 11.84CCDD1338 pKa = 3.3RR1339 pKa = 11.84YY1340 pKa = 10.78EE1341 pKa = 4.28EE1342 pKa = 4.54YY1343 pKa = 10.79DD1344 pKa = 3.78LEE1346 pKa = 4.13PHH1348 pKa = 6.39KK1349 pKa = 11.21VHH1351 pKa = 6.25VHH1353 pKa = 5.33
Molecular weight: 149.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T2BBK0|T2BBK0_MERS Nucleoprotein OS=Middle East respiratory syndrome-related coronavirus OX=1335626 GN=N PE=3 SV=1
MM1 pKa = 7.86ASPAAPRR8 pKa = 11.84AVSFADD14 pKa = 3.78NNDD17 pKa = 3.02ITNTNLSRR25 pKa = 11.84GRR27 pKa = 11.84GRR29 pKa = 11.84NPKK32 pKa = 9.26PRR34 pKa = 11.84AAPNNTVSWYY44 pKa = 9.65TGLTQHH50 pKa = 6.38GKK52 pKa = 10.01VPLTFPPGQGVPLNANSTPAQNAGYY77 pKa = 9.33WRR79 pKa = 11.84RR80 pKa = 11.84QDD82 pKa = 3.96RR83 pKa = 11.84KK84 pKa = 9.7INTGNGIKK92 pKa = 9.96QLAPRR97 pKa = 11.84WYY99 pKa = 9.84FYY101 pKa = 10.45YY102 pKa = 10.42TGTGPEE108 pKa = 3.59AALPFRR114 pKa = 11.84AVKK117 pKa = 10.58DD118 pKa = 4.02GIVWVHH124 pKa = 5.14EE125 pKa = 4.51HH126 pKa = 6.58GATDD130 pKa = 3.5APSTFGTRR138 pKa = 11.84NPNNDD143 pKa = 3.31SAIVTQFAPGTKK155 pKa = 9.59LPKK158 pKa = 9.89NFHH161 pKa = 6.42IEE163 pKa = 4.18GTGGNSQSSSRR174 pKa = 11.84ASSVSRR180 pKa = 11.84NSSRR184 pKa = 11.84SSSQGSRR191 pKa = 11.84SGNSTRR197 pKa = 11.84GTSPGPSGIGAVGGDD212 pKa = 4.31LLYY215 pKa = 11.06LDD217 pKa = 5.07LLNRR221 pKa = 11.84LQALEE226 pKa = 4.11SGKK229 pKa = 10.41VKK231 pKa = 10.49QSQPKK236 pKa = 10.37VITKK240 pKa = 10.17KK241 pKa = 10.58DD242 pKa = 2.86AAAAKK247 pKa = 10.32NKK249 pKa = 8.51MRR251 pKa = 11.84HH252 pKa = 5.56KK253 pKa = 9.8RR254 pKa = 11.84TSTKK258 pKa = 10.27SFNMVQAFGLRR269 pKa = 11.84GPGDD273 pKa = 3.51LQGNFGDD280 pKa = 4.05LQLNKK285 pKa = 10.39LGTEE289 pKa = 4.19DD290 pKa = 4.25PRR292 pKa = 11.84WPQIAEE298 pKa = 4.14LAPTASAFMGMSQFKK313 pKa = 8.94LTHH316 pKa = 5.96QNNDD320 pKa = 2.91DD321 pKa = 4.12HH322 pKa = 6.97GNPVYY327 pKa = 10.43FLRR330 pKa = 11.84YY331 pKa = 9.33SGAIKK336 pKa = 10.61LDD338 pKa = 3.71PKK340 pKa = 10.57NPNYY344 pKa = 10.63NKK346 pKa = 9.76WLEE349 pKa = 3.97LLEE352 pKa = 4.17QNIDD356 pKa = 3.26AYY358 pKa = 9.71KK359 pKa = 10.18TFPKK363 pKa = 10.2KK364 pKa = 9.89EE365 pKa = 4.15KK366 pKa = 9.67KK367 pKa = 9.72QKK369 pKa = 10.33APKK372 pKa = 10.05EE373 pKa = 3.99EE374 pKa = 4.54STDD377 pKa = 3.68QMSEE381 pKa = 3.88PPKK384 pKa = 8.43EE385 pKa = 3.81QRR387 pKa = 11.84VQGSITQRR395 pKa = 11.84TRR397 pKa = 11.84TRR399 pKa = 11.84PSVQPGPMIDD409 pKa = 3.5VNTDD413 pKa = 2.58
MM1 pKa = 7.86ASPAAPRR8 pKa = 11.84AVSFADD14 pKa = 3.78NNDD17 pKa = 3.02ITNTNLSRR25 pKa = 11.84GRR27 pKa = 11.84GRR29 pKa = 11.84NPKK32 pKa = 9.26PRR34 pKa = 11.84AAPNNTVSWYY44 pKa = 9.65TGLTQHH50 pKa = 6.38GKK52 pKa = 10.01VPLTFPPGQGVPLNANSTPAQNAGYY77 pKa = 9.33WRR79 pKa = 11.84RR80 pKa = 11.84QDD82 pKa = 3.96RR83 pKa = 11.84KK84 pKa = 9.7INTGNGIKK92 pKa = 9.96QLAPRR97 pKa = 11.84WYY99 pKa = 9.84FYY101 pKa = 10.45YY102 pKa = 10.42TGTGPEE108 pKa = 3.59AALPFRR114 pKa = 11.84AVKK117 pKa = 10.58DD118 pKa = 4.02GIVWVHH124 pKa = 5.14EE125 pKa = 4.51HH126 pKa = 6.58GATDD130 pKa = 3.5APSTFGTRR138 pKa = 11.84NPNNDD143 pKa = 3.31SAIVTQFAPGTKK155 pKa = 9.59LPKK158 pKa = 9.89NFHH161 pKa = 6.42IEE163 pKa = 4.18GTGGNSQSSSRR174 pKa = 11.84ASSVSRR180 pKa = 11.84NSSRR184 pKa = 11.84SSSQGSRR191 pKa = 11.84SGNSTRR197 pKa = 11.84GTSPGPSGIGAVGGDD212 pKa = 4.31LLYY215 pKa = 11.06LDD217 pKa = 5.07LLNRR221 pKa = 11.84LQALEE226 pKa = 4.11SGKK229 pKa = 10.41VKK231 pKa = 10.49QSQPKK236 pKa = 10.37VITKK240 pKa = 10.17KK241 pKa = 10.58DD242 pKa = 2.86AAAAKK247 pKa = 10.32NKK249 pKa = 8.51MRR251 pKa = 11.84HH252 pKa = 5.56KK253 pKa = 9.8RR254 pKa = 11.84TSTKK258 pKa = 10.27SFNMVQAFGLRR269 pKa = 11.84GPGDD273 pKa = 3.51LQGNFGDD280 pKa = 4.05LQLNKK285 pKa = 10.39LGTEE289 pKa = 4.19DD290 pKa = 4.25PRR292 pKa = 11.84WPQIAEE298 pKa = 4.14LAPTASAFMGMSQFKK313 pKa = 8.94LTHH316 pKa = 5.96QNNDD320 pKa = 2.91DD321 pKa = 4.12HH322 pKa = 6.97GNPVYY327 pKa = 10.43FLRR330 pKa = 11.84YY331 pKa = 9.33SGAIKK336 pKa = 10.61LDD338 pKa = 3.71PKK340 pKa = 10.57NPNYY344 pKa = 10.63NKK346 pKa = 9.76WLEE349 pKa = 3.97LLEE352 pKa = 4.17QNIDD356 pKa = 3.26AYY358 pKa = 9.71KK359 pKa = 10.18TFPKK363 pKa = 10.2KK364 pKa = 9.89EE365 pKa = 4.15KK366 pKa = 9.67KK367 pKa = 9.72QKK369 pKa = 10.33APKK372 pKa = 10.05EE373 pKa = 3.99EE374 pKa = 4.54STDD377 pKa = 3.68QMSEE381 pKa = 3.88PPKK384 pKa = 8.43EE385 pKa = 3.81QRR387 pKa = 11.84VQGSITQRR395 pKa = 11.84TRR397 pKa = 11.84TRR399 pKa = 11.84PSVQPGPMIDD409 pKa = 3.5VNTDD413 pKa = 2.58
Molecular weight: 45.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14330 |
82 |
7078 |
1302.7 |
144.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.264 ± 0.276 | 3.001 ± 0.308 |
5.304 ± 0.332 | 3.88 ± 0.195 |
5.045 ± 0.17 | 5.827 ± 0.241 |
2.066 ± 0.172 | 4.801 ± 0.183 |
5.255 ± 0.352 | 9.463 ± 0.382 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.247 ± 0.148 | 4.997 ± 0.161 |
4.215 ± 0.459 | 3.489 ± 0.301 |
3.643 ± 0.292 | 7.704 ± 0.511 |
6.936 ± 0.1 | 9.016 ± 0.587 |
1.137 ± 0.089 | 4.71 ± 0.242 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
