
Orf virus (strain Goat/Texas/SA00/2000) (OV-SA00) (Orf virus-San Angelo 2000)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Parapoxvirus; Orf virus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
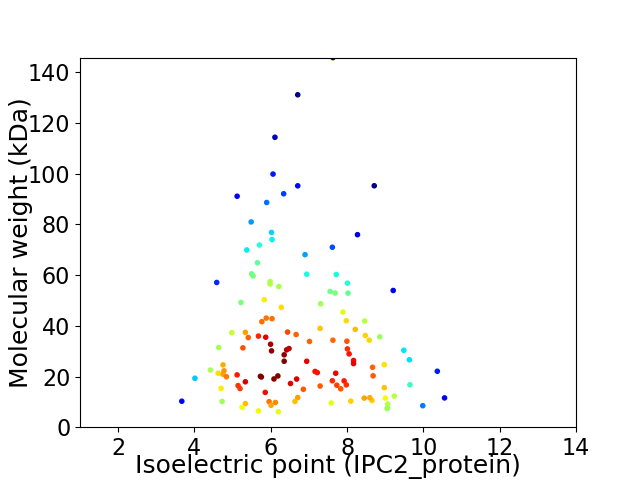
Virtual 2D-PAGE plot for 129 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6TVP1|Q6TVP1_ORFSA Major core protein 4b OS=Orf virus (strain Goat/Texas/SA00/2000) OX=647330 PE=3 SV=1
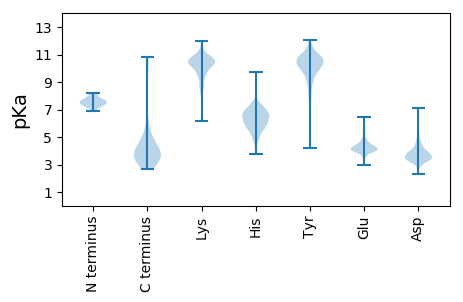
MM1 pKa = 7.37EE2 pKa = 4.76EE3 pKa = 4.16SVAVEE8 pKa = 3.98YY9 pKa = 11.02ADD11 pKa = 3.81EE12 pKa = 5.79DD13 pKa = 3.71EE14 pKa = 5.97DD15 pKa = 4.6EE16 pKa = 5.02IEE18 pKa = 4.13EE19 pKa = 4.35EE20 pKa = 4.33YY21 pKa = 11.11EE22 pKa = 4.32EE23 pKa = 4.32EE24 pKa = 5.49DD25 pKa = 3.33EE26 pKa = 5.62DD27 pKa = 4.63EE28 pKa = 4.6EE29 pKa = 4.59EE30 pKa = 4.35EE31 pKa = 4.42SAEE34 pKa = 4.07GAADD38 pKa = 3.62SSVSDD43 pKa = 3.77VALSAAEE50 pKa = 4.02KK51 pKa = 10.32LVASEE56 pKa = 4.66VPDD59 pKa = 4.1DD60 pKa = 4.48AAAADD65 pKa = 3.83TNVRR69 pKa = 11.84QRR71 pKa = 11.84VTARR75 pKa = 11.84VEE77 pKa = 3.97EE78 pKa = 4.33LKK80 pKa = 10.9ARR82 pKa = 11.84YY83 pKa = 6.43TRR85 pKa = 11.84RR86 pKa = 11.84MSLFEE91 pKa = 3.78LTGIVAEE98 pKa = 4.73SFNLLCRR105 pKa = 11.84GRR107 pKa = 11.84LPLVADD113 pKa = 4.18AADD116 pKa = 3.69PALDD120 pKa = 3.83NEE122 pKa = 4.61LKK124 pKa = 10.61VVVRR128 pKa = 11.84EE129 pKa = 4.22LEE131 pKa = 4.08EE132 pKa = 4.58GVCPIVIEE140 pKa = 4.32KK141 pKa = 10.61NGEE144 pKa = 3.89FLSPGDD150 pKa = 4.39FDD152 pKa = 4.67PEE154 pKa = 4.04CLRR157 pKa = 11.84YY158 pKa = 9.61HH159 pKa = 6.41LTYY162 pKa = 8.24MTDD165 pKa = 2.63LWKK168 pKa = 11.0SQGRR172 pKa = 11.84MM173 pKa = 3.26
MM1 pKa = 7.37EE2 pKa = 4.76EE3 pKa = 4.16SVAVEE8 pKa = 3.98YY9 pKa = 11.02ADD11 pKa = 3.81EE12 pKa = 5.79DD13 pKa = 3.71EE14 pKa = 5.97DD15 pKa = 4.6EE16 pKa = 5.02IEE18 pKa = 4.13EE19 pKa = 4.35EE20 pKa = 4.33YY21 pKa = 11.11EE22 pKa = 4.32EE23 pKa = 4.32EE24 pKa = 5.49DD25 pKa = 3.33EE26 pKa = 5.62DD27 pKa = 4.63EE28 pKa = 4.6EE29 pKa = 4.59EE30 pKa = 4.35EE31 pKa = 4.42SAEE34 pKa = 4.07GAADD38 pKa = 3.62SSVSDD43 pKa = 3.77VALSAAEE50 pKa = 4.02KK51 pKa = 10.32LVASEE56 pKa = 4.66VPDD59 pKa = 4.1DD60 pKa = 4.48AAAADD65 pKa = 3.83TNVRR69 pKa = 11.84QRR71 pKa = 11.84VTARR75 pKa = 11.84VEE77 pKa = 3.97EE78 pKa = 4.33LKK80 pKa = 10.9ARR82 pKa = 11.84YY83 pKa = 6.43TRR85 pKa = 11.84RR86 pKa = 11.84MSLFEE91 pKa = 3.78LTGIVAEE98 pKa = 4.73SFNLLCRR105 pKa = 11.84GRR107 pKa = 11.84LPLVADD113 pKa = 4.18AADD116 pKa = 3.69PALDD120 pKa = 3.83NEE122 pKa = 4.61LKK124 pKa = 10.61VVVRR128 pKa = 11.84EE129 pKa = 4.22LEE131 pKa = 4.08EE132 pKa = 4.58GVCPIVIEE140 pKa = 4.32KK141 pKa = 10.61NGEE144 pKa = 3.89FLSPGDD150 pKa = 4.39FDD152 pKa = 4.67PEE154 pKa = 4.04CLRR157 pKa = 11.84YY158 pKa = 9.61HH159 pKa = 6.41LTYY162 pKa = 8.24MTDD165 pKa = 2.63LWKK168 pKa = 11.0SQGRR172 pKa = 11.84MM173 pKa = 3.26
Molecular weight: 19.31 kDa
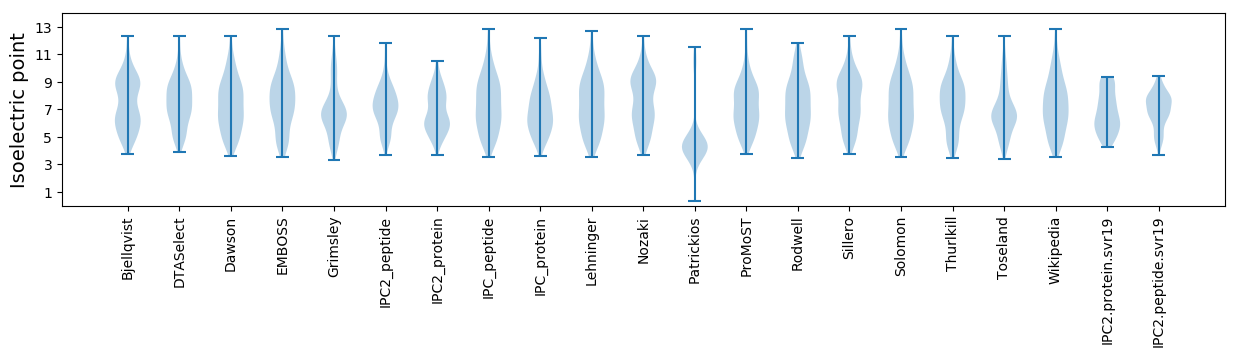
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6TVN4|Q6TVN4_ORFSA Major core protein 4a precursor OS=Orf virus (strain Goat/Texas/SA00/2000) OX=647330 PE=3 SV=1
MM1 pKa = 7.63ASEE4 pKa = 4.98KK5 pKa = 9.75MAQRR9 pKa = 11.84PQSSYY14 pKa = 11.53DD15 pKa = 3.53DD16 pKa = 3.81YY17 pKa = 12.08VNTLQKK23 pKa = 10.13IAPHH27 pKa = 5.86LRR29 pKa = 11.84SLLTQLNNEE38 pKa = 3.88QVIRR42 pKa = 11.84GGATSGDD49 pKa = 3.68VDD51 pKa = 4.08CVGGAGALEE60 pKa = 4.54ASSSSTRR67 pKa = 11.84RR68 pKa = 11.84GSSSSSSRR76 pKa = 11.84KK77 pKa = 8.14RR78 pKa = 11.84TTGTRR83 pKa = 11.84STGSRR88 pKa = 11.84STGSRR93 pKa = 11.84SSGTRR98 pKa = 11.84SSSSTRR104 pKa = 11.84SRR106 pKa = 11.84RR107 pKa = 11.84SGAPSRR113 pKa = 11.84RR114 pKa = 11.84NNDD117 pKa = 2.69GVYY120 pKa = 10.6SGMNACDD127 pKa = 4.55DD128 pKa = 3.9EE129 pKa = 4.69TTIQAVSNCGKK140 pKa = 9.93IVYY143 pKa = 9.81GVVKK147 pKa = 10.54NGKK150 pKa = 9.45LDD152 pKa = 3.46VQGTVGEE159 pKa = 4.35VCEE162 pKa = 4.78DD163 pKa = 3.54LLGIDD168 pKa = 4.0SVNGGRR174 pKa = 11.84KK175 pKa = 9.2KK176 pKa = 10.72SSSGKK181 pKa = 10.21SGTGTSRR188 pKa = 11.84RR189 pKa = 11.84GGTGTGTGRR198 pKa = 11.84SSTSGTTGRR207 pKa = 11.84RR208 pKa = 11.84STNSGTSSSASSSRR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84ASSSNTPCSTTSSASTGARR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84ASNLMNAMDD254 pKa = 5.46LDD256 pKa = 4.16SGMCC260 pKa = 3.85
MM1 pKa = 7.63ASEE4 pKa = 4.98KK5 pKa = 9.75MAQRR9 pKa = 11.84PQSSYY14 pKa = 11.53DD15 pKa = 3.53DD16 pKa = 3.81YY17 pKa = 12.08VNTLQKK23 pKa = 10.13IAPHH27 pKa = 5.86LRR29 pKa = 11.84SLLTQLNNEE38 pKa = 3.88QVIRR42 pKa = 11.84GGATSGDD49 pKa = 3.68VDD51 pKa = 4.08CVGGAGALEE60 pKa = 4.54ASSSSTRR67 pKa = 11.84RR68 pKa = 11.84GSSSSSSRR76 pKa = 11.84KK77 pKa = 8.14RR78 pKa = 11.84TTGTRR83 pKa = 11.84STGSRR88 pKa = 11.84STGSRR93 pKa = 11.84SSGTRR98 pKa = 11.84SSSSTRR104 pKa = 11.84SRR106 pKa = 11.84RR107 pKa = 11.84SGAPSRR113 pKa = 11.84RR114 pKa = 11.84NNDD117 pKa = 2.69GVYY120 pKa = 10.6SGMNACDD127 pKa = 4.55DD128 pKa = 3.9EE129 pKa = 4.69TTIQAVSNCGKK140 pKa = 9.93IVYY143 pKa = 9.81GVVKK147 pKa = 10.54NGKK150 pKa = 9.45LDD152 pKa = 3.46VQGTVGEE159 pKa = 4.35VCEE162 pKa = 4.78DD163 pKa = 3.54LLGIDD168 pKa = 4.0SVNGGRR174 pKa = 11.84KK175 pKa = 9.2KK176 pKa = 10.72SSSGKK181 pKa = 10.21SGTGTSRR188 pKa = 11.84RR189 pKa = 11.84GGTGTGTGRR198 pKa = 11.84SSTSGTTGRR207 pKa = 11.84RR208 pKa = 11.84STNSGTSSSASSSRR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84ASSSNTPCSTTSSASTGARR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84ASNLMNAMDD254 pKa = 5.46LDD256 pKa = 4.16SGMCC260 pKa = 3.85
Molecular weight: 26.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
42036 |
53 |
1289 |
325.9 |
36.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.379 ± 0.315 | 2.391 ± 0.119 |
5.916 ± 0.117 | 5.885 ± 0.128 |
4.527 ± 0.164 | 5.486 ± 0.15 |
2.177 ± 0.096 | 3.778 ± 0.159 |
3.59 ± 0.164 | 9.585 ± 0.199 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.829 ± 0.107 | 3.454 ± 0.144 |
4.989 ± 0.224 | 2.129 ± 0.082 |
8.2 ± 0.212 | 6.98 ± 0.215 |
5.374 ± 0.155 | 8.757 ± 0.172 |
0.692 ± 0.057 | 2.883 ± 0.114 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
