
Palleronia marisminoris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Palleronia
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
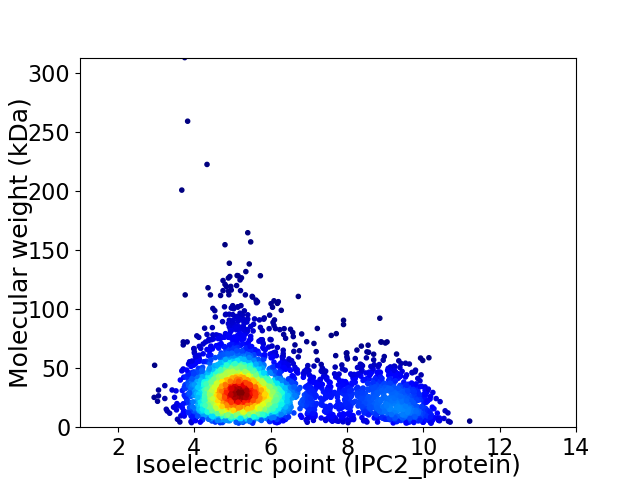
Virtual 2D-PAGE plot for 3664 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y5THL1|A0A1Y5THL1_9RHOB Aminoglycoside N(3)-acetyltransferase OS=Palleronia marisminoris OX=315423 GN=yokD PE=3 SV=1
MM1 pKa = 7.16TFKK4 pKa = 8.65TTLYY8 pKa = 9.64TGAAMVALGTGMAQAQDD25 pKa = 3.61FVFPKK30 pKa = 10.97GEE32 pKa = 4.32GAFSWDD38 pKa = 3.4SLEE41 pKa = 4.5EE42 pKa = 3.82FASNHH47 pKa = 6.71DD48 pKa = 3.86YY49 pKa = 11.06SGQSIDD55 pKa = 3.99ITGPWTGADD64 pKa = 3.57AEE66 pKa = 4.33LMEE69 pKa = 4.69SVVAYY74 pKa = 10.21FEE76 pKa = 4.4EE77 pKa = 4.49ATGASVNYY85 pKa = 9.88SGSDD89 pKa = 3.11SFEE92 pKa = 3.74QDD94 pKa = 2.53IVISTEE100 pKa = 3.85ANSAPNIAVFPQPGLLADD118 pKa = 4.5LAQRR122 pKa = 11.84GAITPLEE129 pKa = 4.47PEE131 pKa = 3.62TAEE134 pKa = 4.25WIRR137 pKa = 11.84EE138 pKa = 4.02NYY140 pKa = 9.31SAGEE144 pKa = 4.11SWVDD148 pKa = 3.31LASFEE153 pKa = 4.5GEE155 pKa = 3.86GGEE158 pKa = 3.94EE159 pKa = 3.65HH160 pKa = 7.24LYY162 pKa = 10.93AFPYY166 pKa = 10.05KK167 pKa = 9.8IDD169 pKa = 3.59VKK171 pKa = 10.85SLVWYY176 pKa = 10.29VPEE179 pKa = 3.88QFEE182 pKa = 4.18EE183 pKa = 4.0AGYY186 pKa = 9.5EE187 pKa = 4.09VPEE190 pKa = 4.31TYY192 pKa = 10.29EE193 pKa = 3.96GLKK196 pKa = 10.62EE197 pKa = 4.02LTQQMADD204 pKa = 3.58DD205 pKa = 6.02GVTPWCIGLGSGAATGWPATDD226 pKa = 3.12WVEE229 pKa = 4.87DD230 pKa = 3.63LMLRR234 pKa = 11.84TATPEE239 pKa = 3.91QYY241 pKa = 10.55DD242 pKa = 3.14AWVANDD248 pKa = 4.15LAFDD252 pKa = 3.74SPEE255 pKa = 3.76VVNAIEE261 pKa = 4.26EE262 pKa = 4.48YY263 pKa = 11.24GFFLQDD269 pKa = 3.11GFVNGGRR276 pKa = 11.84EE277 pKa = 3.91AAATTDD283 pKa = 4.18FRR285 pKa = 11.84DD286 pKa = 3.76SPSGLFQFPPEE297 pKa = 4.47CYY299 pKa = 8.14MHH301 pKa = 6.63KK302 pKa = 10.01QATFIPTFFPEE313 pKa = 4.54GSEE316 pKa = 4.0VGTDD320 pKa = 2.69VDD322 pKa = 4.83FFYY325 pKa = 11.09FPAPEE330 pKa = 5.09GGDD333 pKa = 3.25AEE335 pKa = 4.41QPVLGAGTMFAITNDD350 pKa = 3.56SEE352 pKa = 4.2PARR355 pKa = 11.84GFIEE359 pKa = 4.23YY360 pKa = 10.11LQTPLAHH367 pKa = 6.86EE368 pKa = 4.22IWAAQGGLLTPHH380 pKa = 6.75TGINPEE386 pKa = 4.0VFGTDD391 pKa = 3.29DD392 pKa = 3.5QRR394 pKa = 11.84ALNDD398 pKa = 3.39ILLDD402 pKa = 3.47ATTFRR407 pKa = 11.84FDD409 pKa = 5.22ASDD412 pKa = 3.81LMPGEE417 pKa = 4.37IGAGAFWTQMVEE429 pKa = 4.28FTTGSQDD436 pKa = 3.18AQATGEE442 pKa = 4.5AIQSRR447 pKa = 11.84WDD449 pKa = 3.34QLSNN453 pKa = 3.42
MM1 pKa = 7.16TFKK4 pKa = 8.65TTLYY8 pKa = 9.64TGAAMVALGTGMAQAQDD25 pKa = 3.61FVFPKK30 pKa = 10.97GEE32 pKa = 4.32GAFSWDD38 pKa = 3.4SLEE41 pKa = 4.5EE42 pKa = 3.82FASNHH47 pKa = 6.71DD48 pKa = 3.86YY49 pKa = 11.06SGQSIDD55 pKa = 3.99ITGPWTGADD64 pKa = 3.57AEE66 pKa = 4.33LMEE69 pKa = 4.69SVVAYY74 pKa = 10.21FEE76 pKa = 4.4EE77 pKa = 4.49ATGASVNYY85 pKa = 9.88SGSDD89 pKa = 3.11SFEE92 pKa = 3.74QDD94 pKa = 2.53IVISTEE100 pKa = 3.85ANSAPNIAVFPQPGLLADD118 pKa = 4.5LAQRR122 pKa = 11.84GAITPLEE129 pKa = 4.47PEE131 pKa = 3.62TAEE134 pKa = 4.25WIRR137 pKa = 11.84EE138 pKa = 4.02NYY140 pKa = 9.31SAGEE144 pKa = 4.11SWVDD148 pKa = 3.31LASFEE153 pKa = 4.5GEE155 pKa = 3.86GGEE158 pKa = 3.94EE159 pKa = 3.65HH160 pKa = 7.24LYY162 pKa = 10.93AFPYY166 pKa = 10.05KK167 pKa = 9.8IDD169 pKa = 3.59VKK171 pKa = 10.85SLVWYY176 pKa = 10.29VPEE179 pKa = 3.88QFEE182 pKa = 4.18EE183 pKa = 4.0AGYY186 pKa = 9.5EE187 pKa = 4.09VPEE190 pKa = 4.31TYY192 pKa = 10.29EE193 pKa = 3.96GLKK196 pKa = 10.62EE197 pKa = 4.02LTQQMADD204 pKa = 3.58DD205 pKa = 6.02GVTPWCIGLGSGAATGWPATDD226 pKa = 3.12WVEE229 pKa = 4.87DD230 pKa = 3.63LMLRR234 pKa = 11.84TATPEE239 pKa = 3.91QYY241 pKa = 10.55DD242 pKa = 3.14AWVANDD248 pKa = 4.15LAFDD252 pKa = 3.74SPEE255 pKa = 3.76VVNAIEE261 pKa = 4.26EE262 pKa = 4.48YY263 pKa = 11.24GFFLQDD269 pKa = 3.11GFVNGGRR276 pKa = 11.84EE277 pKa = 3.91AAATTDD283 pKa = 4.18FRR285 pKa = 11.84DD286 pKa = 3.76SPSGLFQFPPEE297 pKa = 4.47CYY299 pKa = 8.14MHH301 pKa = 6.63KK302 pKa = 10.01QATFIPTFFPEE313 pKa = 4.54GSEE316 pKa = 4.0VGTDD320 pKa = 2.69VDD322 pKa = 4.83FFYY325 pKa = 11.09FPAPEE330 pKa = 5.09GGDD333 pKa = 3.25AEE335 pKa = 4.41QPVLGAGTMFAITNDD350 pKa = 3.56SEE352 pKa = 4.2PARR355 pKa = 11.84GFIEE359 pKa = 4.23YY360 pKa = 10.11LQTPLAHH367 pKa = 6.86EE368 pKa = 4.22IWAAQGGLLTPHH380 pKa = 6.75TGINPEE386 pKa = 4.0VFGTDD391 pKa = 3.29DD392 pKa = 3.5QRR394 pKa = 11.84ALNDD398 pKa = 3.39ILLDD402 pKa = 3.47ATTFRR407 pKa = 11.84FDD409 pKa = 5.22ASDD412 pKa = 3.81LMPGEE417 pKa = 4.37IGAGAFWTQMVEE429 pKa = 4.28FTTGSQDD436 pKa = 3.18AQATGEE442 pKa = 4.5AIQSRR447 pKa = 11.84WDD449 pKa = 3.34QLSNN453 pKa = 3.42
Molecular weight: 49.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y5T7K7|A0A1Y5T7K7_9RHOB Sec-independent protein translocase protein TatB OS=Palleronia marisminoris OX=315423 GN=tatB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.3KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1119639 |
29 |
3005 |
305.6 |
33.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.471 ± 0.059 | 0.844 ± 0.012 |
6.267 ± 0.042 | 6.232 ± 0.035 |
3.596 ± 0.031 | 8.893 ± 0.039 |
2.053 ± 0.019 | 5.022 ± 0.029 |
2.559 ± 0.033 | 9.994 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.657 ± 0.021 | 2.334 ± 0.025 |
5.25 ± 0.035 | 2.957 ± 0.024 |
7.328 ± 0.048 | 5.063 ± 0.028 |
5.519 ± 0.026 | 7.457 ± 0.033 |
1.401 ± 0.017 | 2.102 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
