
Rice latent virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.96
Get precalculated fractions of proteins
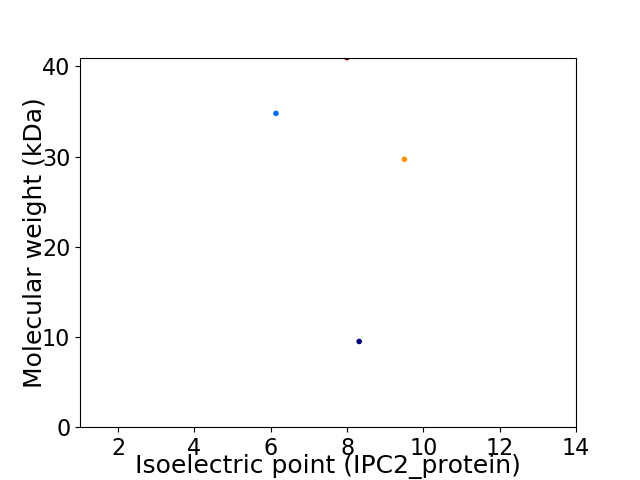
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
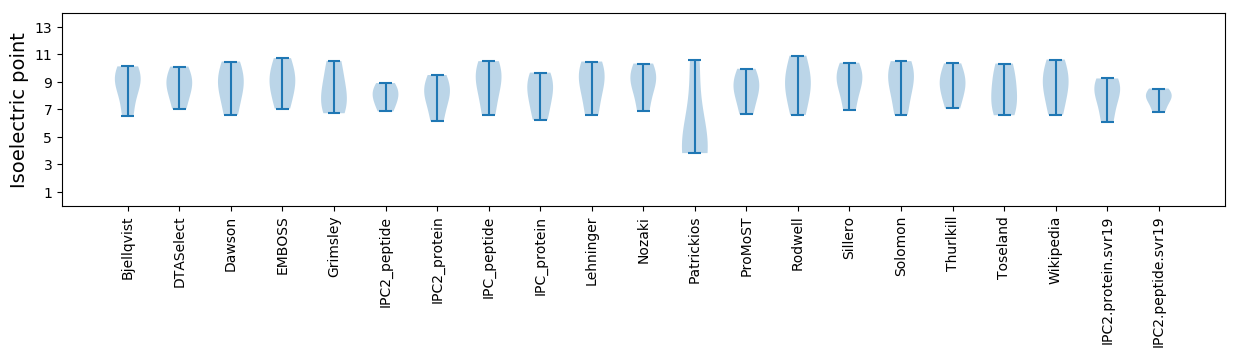
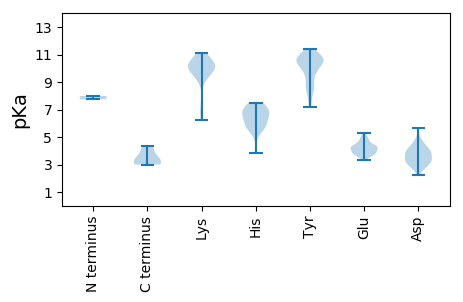
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D0WZA5|A0A2D0WZA5_9GEMI Capsid protein OS=Rice latent virus 1 OX=2012856 PE=3 SV=1
MM1 pKa = 7.99ADD3 pKa = 3.99PSTSHH8 pKa = 5.68QFSVRR13 pKa = 11.84TKK15 pKa = 10.32YY16 pKa = 10.6FFLTYY21 pKa = 8.83PKK23 pKa = 10.48CPVDD27 pKa = 3.32PWEE30 pKa = 4.31LLHH33 pKa = 7.13YY34 pKa = 10.67LLTLLHH40 pKa = 6.54SRR42 pKa = 11.84VPIYY46 pKa = 10.51ILVTRR51 pKa = 11.84EE52 pKa = 3.75SHH54 pKa = 6.98KK55 pKa = 11.13DD56 pKa = 3.41GSFHH60 pKa = 7.46LHH62 pKa = 6.01TLLQLSNRR70 pKa = 11.84LEE72 pKa = 4.37TKK74 pKa = 10.25DD75 pKa = 3.94SRR77 pKa = 11.84FFDD80 pKa = 3.85YY81 pKa = 10.76QSYY84 pKa = 10.5HH85 pKa = 6.91PNCQPAQNCKK95 pKa = 7.13QVRR98 pKa = 11.84DD99 pKa = 4.24YY100 pKa = 9.72ITKK103 pKa = 9.91PGTVSIAEE111 pKa = 3.71WGEE114 pKa = 3.87FYY116 pKa = 10.59NAKK119 pKa = 9.98PFSKK123 pKa = 9.3KK124 pKa = 6.7TLKK127 pKa = 10.33RR128 pKa = 11.84RR129 pKa = 11.84APGSDD134 pKa = 2.84TKK136 pKa = 10.05DD137 pKa = 2.75TKK139 pKa = 9.64MRR141 pKa = 11.84KK142 pKa = 9.06IISSSTSKK150 pKa = 9.8TEE152 pKa = 3.7YY153 pKa = 10.61LSMVRR158 pKa = 11.84QTFPFDD164 pKa = 2.8WATRR168 pKa = 11.84LAQFEE173 pKa = 4.35YY174 pKa = 10.47SAEE177 pKa = 4.09KK178 pKa = 10.68LFPTTVTPYY187 pKa = 10.62ASSVPTDD194 pKa = 3.25QLRR197 pKa = 11.84CSEE200 pKa = 4.84QLSQWATEE208 pKa = 4.17NIKK211 pKa = 9.88TYY213 pKa = 7.19TVCEE217 pKa = 3.85EE218 pKa = 4.96VYY220 pKa = 9.35TQTRR224 pKa = 11.84QNMDD228 pKa = 3.65YY229 pKa = 11.28LDD231 pKa = 3.82VTTAEE236 pKa = 4.52YY237 pKa = 9.99LQHH240 pKa = 6.62VHH242 pKa = 7.37DD243 pKa = 4.46LTSRR247 pKa = 11.84MLQQPPEE254 pKa = 4.88DD255 pKa = 4.0IPPYY259 pKa = 9.79MYY261 pKa = 10.86ADD263 pKa = 3.37QPEE266 pKa = 4.25QEE268 pKa = 4.21RR269 pKa = 11.84QPGQEE274 pKa = 3.85ASGSTTIGTDD284 pKa = 2.26WWTSPLMTPPHH295 pKa = 5.55NTMLL299 pKa = 4.31
MM1 pKa = 7.99ADD3 pKa = 3.99PSTSHH8 pKa = 5.68QFSVRR13 pKa = 11.84TKK15 pKa = 10.32YY16 pKa = 10.6FFLTYY21 pKa = 8.83PKK23 pKa = 10.48CPVDD27 pKa = 3.32PWEE30 pKa = 4.31LLHH33 pKa = 7.13YY34 pKa = 10.67LLTLLHH40 pKa = 6.54SRR42 pKa = 11.84VPIYY46 pKa = 10.51ILVTRR51 pKa = 11.84EE52 pKa = 3.75SHH54 pKa = 6.98KK55 pKa = 11.13DD56 pKa = 3.41GSFHH60 pKa = 7.46LHH62 pKa = 6.01TLLQLSNRR70 pKa = 11.84LEE72 pKa = 4.37TKK74 pKa = 10.25DD75 pKa = 3.94SRR77 pKa = 11.84FFDD80 pKa = 3.85YY81 pKa = 10.76QSYY84 pKa = 10.5HH85 pKa = 6.91PNCQPAQNCKK95 pKa = 7.13QVRR98 pKa = 11.84DD99 pKa = 4.24YY100 pKa = 9.72ITKK103 pKa = 9.91PGTVSIAEE111 pKa = 3.71WGEE114 pKa = 3.87FYY116 pKa = 10.59NAKK119 pKa = 9.98PFSKK123 pKa = 9.3KK124 pKa = 6.7TLKK127 pKa = 10.33RR128 pKa = 11.84RR129 pKa = 11.84APGSDD134 pKa = 2.84TKK136 pKa = 10.05DD137 pKa = 2.75TKK139 pKa = 9.64MRR141 pKa = 11.84KK142 pKa = 9.06IISSSTSKK150 pKa = 9.8TEE152 pKa = 3.7YY153 pKa = 10.61LSMVRR158 pKa = 11.84QTFPFDD164 pKa = 2.8WATRR168 pKa = 11.84LAQFEE173 pKa = 4.35YY174 pKa = 10.47SAEE177 pKa = 4.09KK178 pKa = 10.68LFPTTVTPYY187 pKa = 10.62ASSVPTDD194 pKa = 3.25QLRR197 pKa = 11.84CSEE200 pKa = 4.84QLSQWATEE208 pKa = 4.17NIKK211 pKa = 9.88TYY213 pKa = 7.19TVCEE217 pKa = 3.85EE218 pKa = 4.96VYY220 pKa = 9.35TQTRR224 pKa = 11.84QNMDD228 pKa = 3.65YY229 pKa = 11.28LDD231 pKa = 3.82VTTAEE236 pKa = 4.52YY237 pKa = 9.99LQHH240 pKa = 6.62VHH242 pKa = 7.37DD243 pKa = 4.46LTSRR247 pKa = 11.84MLQQPPEE254 pKa = 4.88DD255 pKa = 4.0IPPYY259 pKa = 9.79MYY261 pKa = 10.86ADD263 pKa = 3.37QPEE266 pKa = 4.25QEE268 pKa = 4.21RR269 pKa = 11.84QPGQEE274 pKa = 3.85ASGSTTIGTDD284 pKa = 2.26WWTSPLMTPPHH295 pKa = 5.55NTMLL299 pKa = 4.31
Molecular weight: 34.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D0WZC9|A0A2D0WZC9_9GEMI Replication-associated protein OS=Rice latent virus 1 OX=2012856 PE=3 SV=1
MM1 pKa = 7.76PGGSKK6 pKa = 10.08RR7 pKa = 11.84ASSGKK12 pKa = 10.13DD13 pKa = 2.87GSRR16 pKa = 11.84PKK18 pKa = 10.08RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.04GEE23 pKa = 3.73PEE25 pKa = 3.33AAFRR29 pKa = 11.84ARR31 pKa = 11.84ILGWASSQFGRR42 pKa = 11.84RR43 pKa = 11.84VYY45 pKa = 10.91GGVGSSQSAPLQVRR59 pKa = 11.84TYY61 pKa = 10.48KK62 pKa = 10.04WEE64 pKa = 4.29NPDD67 pKa = 3.25KK68 pKa = 11.01SAIAIDD74 pKa = 3.84TGSIWIVNTLDD85 pKa = 3.95AGNGDD90 pKa = 4.34DD91 pKa = 4.91QRR93 pKa = 11.84HH94 pKa = 3.87THH96 pKa = 5.3KK97 pKa = 9.91TIIYY101 pKa = 9.83KK102 pKa = 9.32MLFQGTMWINDD113 pKa = 3.42ATSAVCGPLTVHH125 pKa = 6.61FWLVYY130 pKa = 9.55DD131 pKa = 4.63AAPKK135 pKa = 10.52GVLPALTDD143 pKa = 3.18IFEE146 pKa = 4.65EE147 pKa = 5.08GFSGLGSTWVVNRR160 pKa = 11.84DD161 pKa = 3.03NVHH164 pKa = 5.9RR165 pKa = 11.84FVVKK169 pKa = 10.39RR170 pKa = 11.84RR171 pKa = 11.84WQMRR175 pKa = 11.84LMTNGQKK182 pKa = 10.18PYY184 pKa = 8.58YY185 pKa = 8.51TNRR188 pKa = 11.84SGNGDD193 pKa = 3.58PTSVTIRR200 pKa = 11.84DD201 pKa = 3.44FRR203 pKa = 11.84KK204 pKa = 9.6FFTKK208 pKa = 10.36LRR210 pKa = 11.84TATEE214 pKa = 4.26WKK216 pKa = 10.26NSATGGIGDD225 pKa = 3.8IKK227 pKa = 10.77KK228 pKa = 10.49GALYY232 pKa = 10.12LCAMCSQAPEE242 pKa = 4.08GSARR246 pKa = 11.84AINLGVKK253 pKa = 10.21FRR255 pKa = 11.84GQSRR259 pKa = 11.84IYY261 pKa = 9.56FKK263 pKa = 10.81CTGNQQ268 pKa = 3.0
MM1 pKa = 7.76PGGSKK6 pKa = 10.08RR7 pKa = 11.84ASSGKK12 pKa = 10.13DD13 pKa = 2.87GSRR16 pKa = 11.84PKK18 pKa = 10.08RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.04GEE23 pKa = 3.73PEE25 pKa = 3.33AAFRR29 pKa = 11.84ARR31 pKa = 11.84ILGWASSQFGRR42 pKa = 11.84RR43 pKa = 11.84VYY45 pKa = 10.91GGVGSSQSAPLQVRR59 pKa = 11.84TYY61 pKa = 10.48KK62 pKa = 10.04WEE64 pKa = 4.29NPDD67 pKa = 3.25KK68 pKa = 11.01SAIAIDD74 pKa = 3.84TGSIWIVNTLDD85 pKa = 3.95AGNGDD90 pKa = 4.34DD91 pKa = 4.91QRR93 pKa = 11.84HH94 pKa = 3.87THH96 pKa = 5.3KK97 pKa = 9.91TIIYY101 pKa = 9.83KK102 pKa = 9.32MLFQGTMWINDD113 pKa = 3.42ATSAVCGPLTVHH125 pKa = 6.61FWLVYY130 pKa = 9.55DD131 pKa = 4.63AAPKK135 pKa = 10.52GVLPALTDD143 pKa = 3.18IFEE146 pKa = 4.65EE147 pKa = 5.08GFSGLGSTWVVNRR160 pKa = 11.84DD161 pKa = 3.03NVHH164 pKa = 5.9RR165 pKa = 11.84FVVKK169 pKa = 10.39RR170 pKa = 11.84RR171 pKa = 11.84WQMRR175 pKa = 11.84LMTNGQKK182 pKa = 10.18PYY184 pKa = 8.58YY185 pKa = 8.51TNRR188 pKa = 11.84SGNGDD193 pKa = 3.58PTSVTIRR200 pKa = 11.84DD201 pKa = 3.44FRR203 pKa = 11.84KK204 pKa = 9.6FFTKK208 pKa = 10.36LRR210 pKa = 11.84TATEE214 pKa = 4.26WKK216 pKa = 10.26NSATGGIGDD225 pKa = 3.8IKK227 pKa = 10.77KK228 pKa = 10.49GALYY232 pKa = 10.12LCAMCSQAPEE242 pKa = 4.08GSARR246 pKa = 11.84AINLGVKK253 pKa = 10.21FRR255 pKa = 11.84GQSRR259 pKa = 11.84IYY261 pKa = 9.56FKK263 pKa = 10.81CTGNQQ268 pKa = 3.0
Molecular weight: 29.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1009 |
88 |
354 |
252.3 |
28.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.748 ± 0.884 | 1.982 ± 0.344 |
5.055 ± 0.288 | 4.262 ± 0.595 |
4.658 ± 0.544 | 5.847 ± 1.787 |
2.478 ± 0.535 | 4.163 ± 0.492 |
6.046 ± 0.781 | 7.334 ± 0.94 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.883 ± 0.366 | 3.271 ± 0.463 |
6.145 ± 0.687 | 4.658 ± 0.734 |
6.046 ± 0.674 | 8.127 ± 0.314 |
9.514 ± 0.845 | 5.55 ± 1.152 |
2.279 ± 0.278 | 4.955 ± 0.806 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
