
Enterobacter lignolyticus (strain SCF1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Pluralibacter; [Enterobacter] lignolyticus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
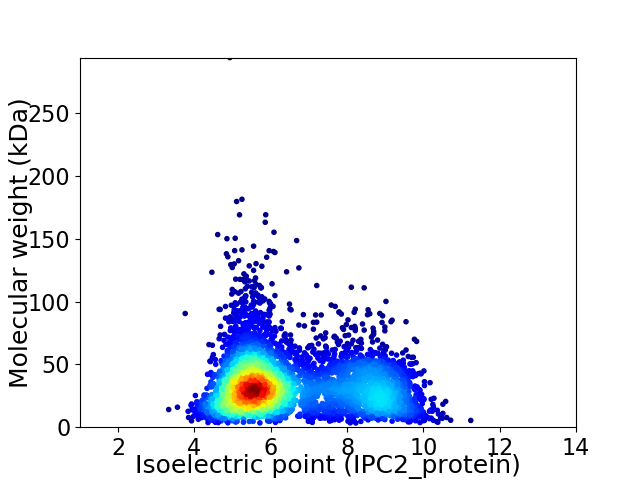
Virtual 2D-PAGE plot for 4393 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E3G2K8|E3G2K8_ENTLS Prophage CP4-57 regulatory OS=Enterobacter lignolyticus (strain SCF1) OX=701347 GN=Entcl_0672 PE=4 SV=1
MM1 pKa = 8.25RR2 pKa = 11.84YY3 pKa = 7.86TSRR6 pKa = 11.84LSLLSHH12 pKa = 7.36CILLSLTSFAVQASATTPSVPCAGGDD38 pKa = 3.27ITQTCGLSVYY48 pKa = 9.65TDD50 pKa = 3.09NNFYY54 pKa = 10.65QGSGDD59 pKa = 3.52KK60 pKa = 10.76DD61 pKa = 3.17AVMAQATATNIYY73 pKa = 9.34MDD75 pKa = 4.04GNRR78 pKa = 11.84RR79 pKa = 11.84AGDD82 pKa = 3.66TQSLIVNGTDD92 pKa = 2.92MSGAYY97 pKa = 8.98IQASKK102 pKa = 11.02GGTVNITMIDD112 pKa = 3.64GASADD117 pKa = 4.05MIEE120 pKa = 5.09SGDD123 pKa = 3.77AGQTTNTAITLDD135 pKa = 3.72DD136 pKa = 4.01AVLKK140 pKa = 11.34GEE142 pKa = 5.03DD143 pKa = 3.87DD144 pKa = 4.3SIAYY148 pKa = 8.41TPGDD152 pKa = 3.45AAGDD156 pKa = 3.75KK157 pKa = 10.69AYY159 pKa = 10.68MMGSAIFIDD168 pKa = 4.38PLDD171 pKa = 4.36AGNHH175 pKa = 4.91SVAITDD181 pKa = 3.7GSLVQGSIVSAGAGVQTIALNNSSLVNGGIYY212 pKa = 9.81TGSNASATTIALTNSVLDD230 pKa = 4.01ASQSEE235 pKa = 4.57IAQQAGEE242 pKa = 4.18IVAGLNEE249 pKa = 4.38KK250 pKa = 10.65GVGPFSNINVQEE262 pKa = 3.99YY263 pKa = 10.39DD264 pKa = 3.5DD265 pKa = 4.42VGIAVYY271 pKa = 9.99GKK273 pKa = 10.14SAMALSLNNSTLTGDD288 pKa = 3.16IALLNEE294 pKa = 4.67GNDD297 pKa = 3.77AGPGSISVTMANNTTVNGDD316 pKa = 3.4MTLVGQSSNTITLTDD331 pKa = 3.99DD332 pKa = 3.18STLNGDD338 pKa = 2.97VDD340 pKa = 3.84AANNSGNTALTLDD353 pKa = 3.64NYY355 pKa = 11.3SRR357 pKa = 11.84INGNIALGSGNDD369 pKa = 3.95TVTLTHH375 pKa = 7.29DD376 pKa = 3.58SQVNGDD382 pKa = 3.51VDD384 pKa = 4.31AGAGDD389 pKa = 4.18DD390 pKa = 4.11TLSMDD395 pKa = 4.02EE396 pKa = 4.62GSTISGQISDD406 pKa = 4.43FEE408 pKa = 4.71TVNTAGNNNIKK419 pKa = 9.57TDD421 pKa = 4.68AINDD425 pKa = 3.64TSTWNLQNGSQLTAQEE441 pKa = 4.34TGSNVQVNMSADD453 pKa = 3.58SYY455 pKa = 11.47VDD457 pKa = 3.58LGTVTGTNDD466 pKa = 3.65EE467 pKa = 4.25IVVNSVTAASVNQQDD482 pKa = 3.76QEE484 pKa = 4.14IGTFTTAGDD493 pKa = 4.09APQTAAAAAFSNGEE507 pKa = 3.86QQVEE511 pKa = 4.53SRR513 pKa = 11.84SGAYY517 pKa = 9.86NYY519 pKa = 10.84SNSLDD524 pKa = 3.54VVAAQSALLTASDD537 pKa = 3.8TQSWDD542 pKa = 3.08ILFSSSRR549 pKa = 11.84SSLASDD555 pKa = 3.71VQGLMAGLDD564 pKa = 3.49AAEE567 pKa = 4.18QAGQLITDD575 pKa = 5.52DD576 pKa = 4.21IANHH580 pKa = 5.12MTQIHH585 pKa = 5.76LQNLRR590 pKa = 11.84GHH592 pKa = 5.58VVGGAQLWGDD602 pKa = 3.72FLYY605 pKa = 11.16QNGDD609 pKa = 3.02VSDD612 pKa = 4.23DD613 pKa = 3.18VDD615 pKa = 4.09YY616 pKa = 11.95NSITQGAQGGVDD628 pKa = 3.2WTIPLANGDD637 pKa = 4.11SLTAGMALGWTRR649 pKa = 11.84SRR651 pKa = 11.84VEE653 pKa = 4.32DD654 pKa = 3.45NSGGEE659 pKa = 4.02NRR661 pKa = 11.84FKK663 pKa = 10.79DD664 pKa = 3.86TVYY667 pKa = 10.78GNYY670 pKa = 9.85YY671 pKa = 10.01SLYY674 pKa = 10.6GGWQQALRR682 pKa = 11.84DD683 pKa = 4.2SRR685 pKa = 11.84WGMFIDD691 pKa = 5.03GSVSYY696 pKa = 11.79GDD698 pKa = 3.39MRR700 pKa = 11.84YY701 pKa = 10.04SLSAQNVTGNTSGMTEE717 pKa = 4.06ALSGSTRR724 pKa = 11.84GDD726 pKa = 3.33LYY728 pKa = 11.07VAQARR733 pKa = 11.84TGVNILLPAEE743 pKa = 4.41TVLQPYY749 pKa = 7.69VTLGWDD755 pKa = 2.83KK756 pKa = 11.28AAADD760 pKa = 5.16GYY762 pKa = 10.87ADD764 pKa = 3.97SEE766 pKa = 4.49IAFSDD771 pKa = 3.59SHH773 pKa = 6.91VSSWNSSVGVHH784 pKa = 6.87LSTTVACLAKK794 pKa = 9.8NTTLTPWLDD803 pKa = 3.16MRR805 pKa = 11.84YY806 pKa = 9.66QGQFSDD812 pKa = 3.7NTDD815 pKa = 2.41IRR817 pKa = 11.84AADD820 pKa = 3.52YY821 pKa = 11.26HH822 pKa = 5.88NTDD825 pKa = 2.76GHH827 pKa = 6.07NLSLGIFGAGISATIAHH844 pKa = 5.89SVALNTGVYY853 pKa = 10.23FGTGDD858 pKa = 3.24VDD860 pKa = 4.08NNASVQAGISYY871 pKa = 10.26RR872 pKa = 11.84FF873 pKa = 3.33
MM1 pKa = 8.25RR2 pKa = 11.84YY3 pKa = 7.86TSRR6 pKa = 11.84LSLLSHH12 pKa = 7.36CILLSLTSFAVQASATTPSVPCAGGDD38 pKa = 3.27ITQTCGLSVYY48 pKa = 9.65TDD50 pKa = 3.09NNFYY54 pKa = 10.65QGSGDD59 pKa = 3.52KK60 pKa = 10.76DD61 pKa = 3.17AVMAQATATNIYY73 pKa = 9.34MDD75 pKa = 4.04GNRR78 pKa = 11.84RR79 pKa = 11.84AGDD82 pKa = 3.66TQSLIVNGTDD92 pKa = 2.92MSGAYY97 pKa = 8.98IQASKK102 pKa = 11.02GGTVNITMIDD112 pKa = 3.64GASADD117 pKa = 4.05MIEE120 pKa = 5.09SGDD123 pKa = 3.77AGQTTNTAITLDD135 pKa = 3.72DD136 pKa = 4.01AVLKK140 pKa = 11.34GEE142 pKa = 5.03DD143 pKa = 3.87DD144 pKa = 4.3SIAYY148 pKa = 8.41TPGDD152 pKa = 3.45AAGDD156 pKa = 3.75KK157 pKa = 10.69AYY159 pKa = 10.68MMGSAIFIDD168 pKa = 4.38PLDD171 pKa = 4.36AGNHH175 pKa = 4.91SVAITDD181 pKa = 3.7GSLVQGSIVSAGAGVQTIALNNSSLVNGGIYY212 pKa = 9.81TGSNASATTIALTNSVLDD230 pKa = 4.01ASQSEE235 pKa = 4.57IAQQAGEE242 pKa = 4.18IVAGLNEE249 pKa = 4.38KK250 pKa = 10.65GVGPFSNINVQEE262 pKa = 3.99YY263 pKa = 10.39DD264 pKa = 3.5DD265 pKa = 4.42VGIAVYY271 pKa = 9.99GKK273 pKa = 10.14SAMALSLNNSTLTGDD288 pKa = 3.16IALLNEE294 pKa = 4.67GNDD297 pKa = 3.77AGPGSISVTMANNTTVNGDD316 pKa = 3.4MTLVGQSSNTITLTDD331 pKa = 3.99DD332 pKa = 3.18STLNGDD338 pKa = 2.97VDD340 pKa = 3.84AANNSGNTALTLDD353 pKa = 3.64NYY355 pKa = 11.3SRR357 pKa = 11.84INGNIALGSGNDD369 pKa = 3.95TVTLTHH375 pKa = 7.29DD376 pKa = 3.58SQVNGDD382 pKa = 3.51VDD384 pKa = 4.31AGAGDD389 pKa = 4.18DD390 pKa = 4.11TLSMDD395 pKa = 4.02EE396 pKa = 4.62GSTISGQISDD406 pKa = 4.43FEE408 pKa = 4.71TVNTAGNNNIKK419 pKa = 9.57TDD421 pKa = 4.68AINDD425 pKa = 3.64TSTWNLQNGSQLTAQEE441 pKa = 4.34TGSNVQVNMSADD453 pKa = 3.58SYY455 pKa = 11.47VDD457 pKa = 3.58LGTVTGTNDD466 pKa = 3.65EE467 pKa = 4.25IVVNSVTAASVNQQDD482 pKa = 3.76QEE484 pKa = 4.14IGTFTTAGDD493 pKa = 4.09APQTAAAAAFSNGEE507 pKa = 3.86QQVEE511 pKa = 4.53SRR513 pKa = 11.84SGAYY517 pKa = 9.86NYY519 pKa = 10.84SNSLDD524 pKa = 3.54VVAAQSALLTASDD537 pKa = 3.8TQSWDD542 pKa = 3.08ILFSSSRR549 pKa = 11.84SSLASDD555 pKa = 3.71VQGLMAGLDD564 pKa = 3.49AAEE567 pKa = 4.18QAGQLITDD575 pKa = 5.52DD576 pKa = 4.21IANHH580 pKa = 5.12MTQIHH585 pKa = 5.76LQNLRR590 pKa = 11.84GHH592 pKa = 5.58VVGGAQLWGDD602 pKa = 3.72FLYY605 pKa = 11.16QNGDD609 pKa = 3.02VSDD612 pKa = 4.23DD613 pKa = 3.18VDD615 pKa = 4.09YY616 pKa = 11.95NSITQGAQGGVDD628 pKa = 3.2WTIPLANGDD637 pKa = 4.11SLTAGMALGWTRR649 pKa = 11.84SRR651 pKa = 11.84VEE653 pKa = 4.32DD654 pKa = 3.45NSGGEE659 pKa = 4.02NRR661 pKa = 11.84FKK663 pKa = 10.79DD664 pKa = 3.86TVYY667 pKa = 10.78GNYY670 pKa = 9.85YY671 pKa = 10.01SLYY674 pKa = 10.6GGWQQALRR682 pKa = 11.84DD683 pKa = 4.2SRR685 pKa = 11.84WGMFIDD691 pKa = 5.03GSVSYY696 pKa = 11.79GDD698 pKa = 3.39MRR700 pKa = 11.84YY701 pKa = 10.04SLSAQNVTGNTSGMTEE717 pKa = 4.06ALSGSTRR724 pKa = 11.84GDD726 pKa = 3.33LYY728 pKa = 11.07VAQARR733 pKa = 11.84TGVNILLPAEE743 pKa = 4.41TVLQPYY749 pKa = 7.69VTLGWDD755 pKa = 2.83KK756 pKa = 11.28AAADD760 pKa = 5.16GYY762 pKa = 10.87ADD764 pKa = 3.97SEE766 pKa = 4.49IAFSDD771 pKa = 3.59SHH773 pKa = 6.91VSSWNSSVGVHH784 pKa = 6.87LSTTVACLAKK794 pKa = 9.8NTTLTPWLDD803 pKa = 3.16MRR805 pKa = 11.84YY806 pKa = 9.66QGQFSDD812 pKa = 3.7NTDD815 pKa = 2.41IRR817 pKa = 11.84AADD820 pKa = 3.52YY821 pKa = 11.26HH822 pKa = 5.88NTDD825 pKa = 2.76GHH827 pKa = 6.07NLSLGIFGAGISATIAHH844 pKa = 5.89SVALNTGVYY853 pKa = 10.23FGTGDD858 pKa = 3.24VDD860 pKa = 4.08NNASVQAGISYY871 pKa = 10.26RR872 pKa = 11.84FF873 pKa = 3.33
Molecular weight: 90.48 kDa
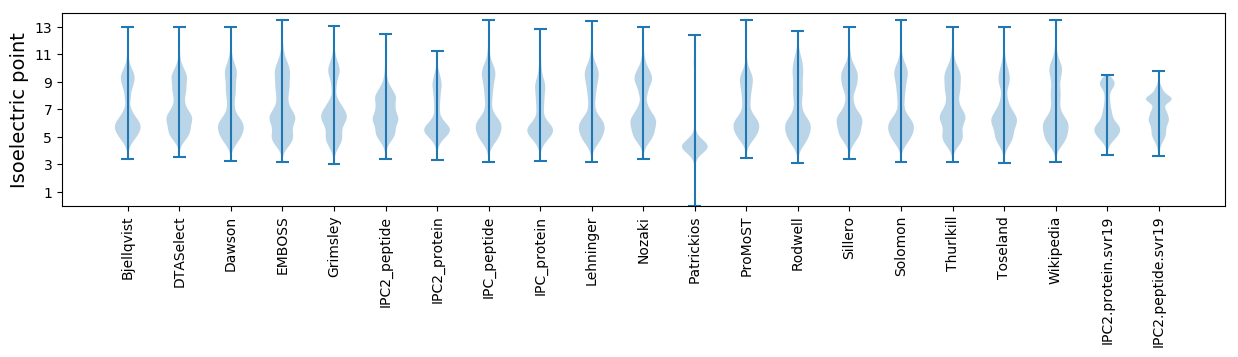
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3G6E6|E3G6E6_ENTLS Thiol:disulfide interchange protein OS=Enterobacter lignolyticus (strain SCF1) OX=701347 GN=Entcl_0997 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1420044 |
30 |
2884 |
323.3 |
35.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.218 ± 0.051 | 1.119 ± 0.013 |
5.206 ± 0.029 | 5.351 ± 0.035 |
3.856 ± 0.024 | 7.59 ± 0.043 |
2.267 ± 0.021 | 5.699 ± 0.031 |
3.962 ± 0.028 | 10.899 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.797 ± 0.016 | 3.621 ± 0.028 |
4.497 ± 0.022 | 4.433 ± 0.029 |
5.787 ± 0.035 | 5.874 ± 0.025 |
5.329 ± 0.023 | 7.212 ± 0.03 |
1.532 ± 0.016 | 2.751 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
