
Pontiella desulfatans
Taxonomy: cellular organisms; Bacteria; PVC group; Kiritimatiellaeota; Kiritimatiellae; Kiritimatiellales; Pontiellaceae; Pontiella
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
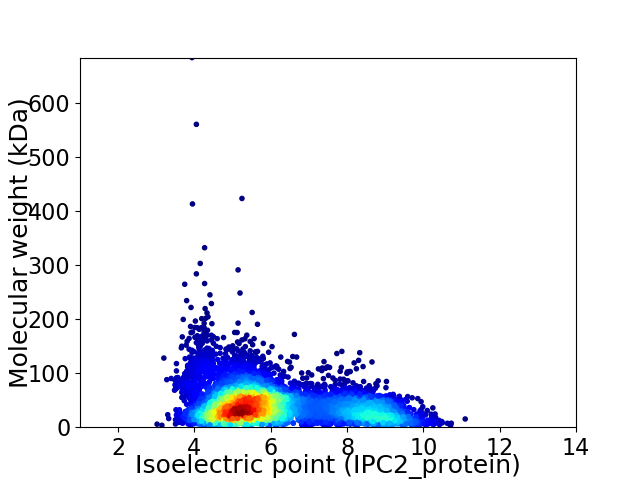
Virtual 2D-PAGE plot for 6374 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
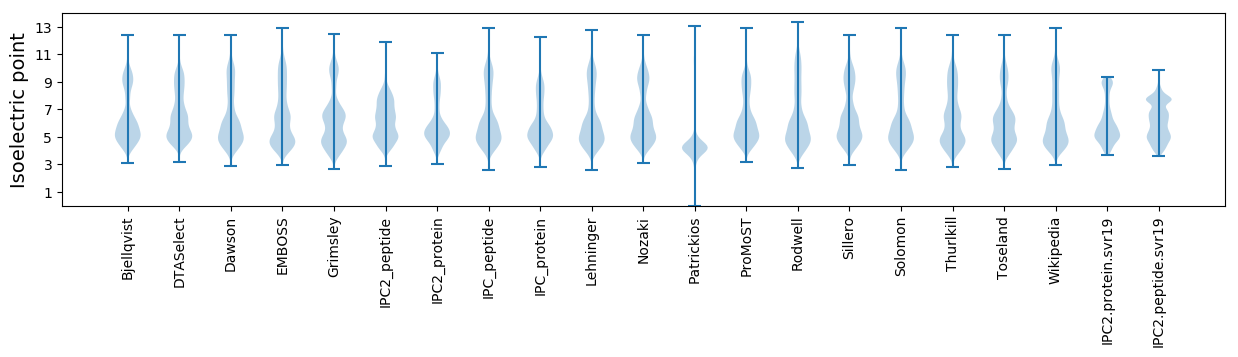
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6C2TX91|A0A6C2TX91_9BACT Signal peptidase I OS=Pontiella desulfatans OX=2750659 GN=sipV PE=3 SV=1
MM1 pKa = 7.29KK2 pKa = 10.27AKK4 pKa = 10.36AAIAGTLLALILAGSAVAQEE24 pKa = 4.37EE25 pKa = 4.52FDD27 pKa = 5.24FGDD30 pKa = 3.97APDD33 pKa = 4.6PGFPTLLANNGARR46 pKa = 11.84HH47 pKa = 5.55LVSIGSQHH55 pKa = 6.3IFLGLAPVDD64 pKa = 4.2TEE66 pKa = 4.75PDD68 pKa = 3.66GQPHH72 pKa = 6.49INAIGDD78 pKa = 4.12DD79 pKa = 3.89NDD81 pKa = 3.46IAYY84 pKa = 7.78PPPFDD89 pKa = 6.06DD90 pKa = 4.31EE91 pKa = 6.47DD92 pKa = 4.13GVSWDD97 pKa = 3.64TPLIPGQVAQITVTASVNGDD117 pKa = 3.04LDD119 pKa = 3.31AWIDD123 pKa = 3.56FSGNNDD129 pKa = 2.69WTTVGDD135 pKa = 4.44QIFTTQPLIAGANTLNFNVPVWASPGYY162 pKa = 7.95TYY164 pKa = 11.55ARR166 pKa = 11.84FRR168 pKa = 11.84FNASGVALQHH178 pKa = 6.2VGPAPDD184 pKa = 4.91GEE186 pKa = 4.42VEE188 pKa = 4.98DD189 pKa = 4.56YY190 pKa = 10.73FIQITQEE197 pKa = 4.14LEE199 pKa = 4.3DD200 pKa = 4.67FYY202 pKa = 11.62DD203 pKa = 3.52WGDD206 pKa = 3.92APDD209 pKa = 3.95SASSPKK215 pKa = 10.43YY216 pKa = 8.36PTLAANNGANHH227 pKa = 7.84LINASVYY234 pKa = 9.96LGAVVDD240 pKa = 4.53IDD242 pKa = 6.08LDD244 pKa = 4.07GQPHH248 pKa = 6.56AKK250 pKa = 10.52ALGDD254 pKa = 5.36DD255 pKa = 3.82INQAYY260 pKa = 8.71PGSPYY265 pKa = 10.42PPGDD269 pKa = 3.3EE270 pKa = 4.96DD271 pKa = 5.5GIAFTSLLVQNQTATLTATASTAGYY296 pKa = 9.56LDD298 pKa = 3.27AWIDD302 pKa = 3.79YY303 pKa = 9.72NQDD306 pKa = 3.53GVWNAAEE313 pKa = 5.04KK314 pKa = 10.41IFSSTPLGAGANALSVTIPGGATAGKK340 pKa = 7.53TFARR344 pKa = 11.84FRR346 pKa = 11.84FSLNGGLAPNGTAPDD361 pKa = 4.29GEE363 pKa = 4.46VEE365 pKa = 4.98DD366 pKa = 4.25YY367 pKa = 10.72EE368 pKa = 4.52VWIEE372 pKa = 4.13TTEE375 pKa = 4.2EE376 pKa = 3.81DD377 pKa = 5.67HH378 pKa = 7.43KK379 pKa = 10.45MHH381 pKa = 6.91WPQYY385 pKa = 8.11PDD387 pKa = 3.08PHH389 pKa = 7.62GWDD392 pKa = 3.17VRR394 pKa = 11.84ASYY397 pKa = 11.45LDD399 pKa = 4.08GNPDD403 pKa = 3.56DD404 pKa = 4.77QFHH407 pKa = 6.84KK408 pKa = 11.09VLADD412 pKa = 4.41DD413 pKa = 5.61FLCTSNGPITKK424 pKa = 8.51ITFWGSFQYY433 pKa = 10.78NGYY436 pKa = 9.96IPTNFHH442 pKa = 6.5KK443 pKa = 10.87FNGISKK449 pKa = 8.89FHH451 pKa = 6.51FSFHH455 pKa = 6.78KK456 pKa = 10.29DD457 pKa = 3.03IPVDD461 pKa = 3.89LPEE464 pKa = 5.4IPYY467 pKa = 10.21SRR469 pKa = 11.84PVVPAEE475 pKa = 3.74WEE477 pKa = 3.82IDD479 pKa = 3.26VDD481 pKa = 4.59PFMLPAGWEE490 pKa = 3.91ITVTTEE496 pKa = 3.58EE497 pKa = 4.76PSWQGWYY504 pKa = 10.25DD505 pKa = 3.94PNTGNWNTNDD515 pKa = 3.11HH516 pKa = 5.31NHH518 pKa = 5.52YY519 pKa = 9.92FRR521 pKa = 11.84YY522 pKa = 10.42DD523 pKa = 3.2ITIPEE528 pKa = 4.28ADD530 pKa = 3.5AFVQVEE536 pKa = 4.4SNIYY540 pKa = 8.89WLDD543 pKa = 3.08ISVDD547 pKa = 3.77LEE549 pKa = 4.12GTEE552 pKa = 6.05GYY554 pKa = 9.15WGWKK558 pKa = 6.85TSRR561 pKa = 11.84SPHH564 pKa = 5.87WNDD567 pKa = 3.1DD568 pKa = 3.97AVWADD573 pKa = 3.86LPVEE577 pKa = 4.59FHH579 pKa = 5.49QQWGEE584 pKa = 4.11LIDD587 pKa = 4.82PIYY590 pKa = 11.19SNSLDD595 pKa = 3.52LAFIINDD602 pKa = 3.34EE603 pKa = 4.31VEE605 pKa = 4.15PEE607 pKa = 3.89EE608 pKa = 4.36TYY610 pKa = 11.3DD611 pKa = 3.56WGDD614 pKa = 4.23APDD617 pKa = 3.7QPYY620 pKa = 7.72PTLAASSGANHH631 pKa = 7.34LIVPGVFMGTLIDD644 pKa = 5.19AEE646 pKa = 4.44TDD648 pKa = 3.53GQPNATATGDD658 pKa = 3.96DD659 pKa = 4.0LAGPAADD666 pKa = 5.3DD667 pKa = 3.55EE668 pKa = 5.24DD669 pKa = 4.24GVVFNTPLFPGQPALIDD686 pKa = 3.6VTVSQAGWLDD696 pKa = 3.14VWLDD700 pKa = 3.34ADD702 pKa = 5.57NNGDD706 pKa = 3.36WLGPNDD712 pKa = 4.57LVYY715 pKa = 10.58SAYY718 pKa = 10.65VSAGLNPLSINIPLSVPPGQNFMRR742 pKa = 11.84FRR744 pKa = 11.84YY745 pKa = 7.45NTNGPLPPTGGAQNGEE761 pKa = 4.18VEE763 pKa = 4.7DD764 pKa = 4.34YY765 pKa = 10.92EE766 pKa = 4.6VDD768 pKa = 3.44IEE770 pKa = 5.4LEE772 pKa = 4.2PEE774 pKa = 4.31LDD776 pKa = 4.62FGDD779 pKa = 4.79APDD782 pKa = 4.55PGYY785 pKa = 8.02PTWLAMNGARR795 pKa = 11.84HH796 pKa = 6.63IIYY799 pKa = 9.96GGPAIALRR807 pKa = 11.84MGAKK811 pKa = 9.66VDD813 pKa = 4.28ADD815 pKa = 3.75PDD817 pKa = 4.11GQPTAASDD825 pKa = 4.03GDD827 pKa = 4.04DD828 pKa = 3.61TDD830 pKa = 5.04GSDD833 pKa = 3.88DD834 pKa = 3.74EE835 pKa = 4.4EE836 pKa = 5.87SIVFTTPIIPGTWAKK851 pKa = 10.01MDD853 pKa = 3.55LTVGVGIPASGYY865 pKa = 10.55AWLQGWADD873 pKa = 3.96FNNDD877 pKa = 4.03GDD879 pKa = 3.91WLDD882 pKa = 3.73AGEE885 pKa = 4.76QIFADD890 pKa = 3.71EE891 pKa = 4.72LVTFGASGSGVNNLSFPVPTTALPGALHH919 pKa = 6.25LRR921 pKa = 11.84LRR923 pKa = 11.84LSTYY927 pKa = 10.2KK928 pKa = 10.93GIGITGLAYY937 pKa = 10.4DD938 pKa = 4.95GEE940 pKa = 4.58VEE942 pKa = 4.44DD943 pKa = 4.36YY944 pKa = 10.34LVHH947 pKa = 7.78VEE949 pKa = 4.33EE950 pKa = 5.41PMDD953 pKa = 4.51IDD955 pKa = 3.84WGDD958 pKa = 4.14AFDD961 pKa = 3.89NWITPVYY968 pKa = 8.71PTILLNNGAHH978 pKa = 5.78HH979 pKa = 6.81VIDD982 pKa = 4.33GVHH985 pKa = 6.16FLGGQIDD992 pKa = 4.16AEE994 pKa = 4.34ADD996 pKa = 3.67GQPAIAVDD1004 pKa = 3.84GDD1006 pKa = 4.73DD1007 pKa = 4.22IVGPSPDD1014 pKa = 4.55DD1015 pKa = 3.44EE1016 pKa = 6.74DD1017 pKa = 5.06GVFFTTMIFAGGYY1030 pKa = 9.45VGVDD1034 pKa = 2.73VDD1036 pKa = 3.7ASTNGVLDD1044 pKa = 3.27AWVDD1048 pKa = 4.04FNNDD1052 pKa = 3.25GDD1054 pKa = 4.02WDD1056 pKa = 4.19DD1057 pKa = 5.15LNEE1060 pKa = 4.8QIFSGQNLHH1069 pKa = 6.97PGINSLGFTVPASAATVYY1087 pKa = 10.48PITSRR1092 pKa = 11.84FRR1094 pKa = 11.84FTSGGISTYY1103 pKa = 10.37TGLAMDD1109 pKa = 5.4GEE1111 pKa = 4.87VEE1113 pKa = 4.35DD1114 pKa = 4.59HH1115 pKa = 7.05ALDD1118 pKa = 3.57IQAVDD1123 pKa = 3.5TNGLDD1128 pKa = 4.33FGDD1131 pKa = 3.96ADD1133 pKa = 3.75IFPEE1137 pKa = 4.38TVLPNGARR1145 pKa = 11.84HH1146 pKa = 6.55AIVPGIVLGSAIDD1159 pKa = 3.81WEE1161 pKa = 4.67PDD1163 pKa = 3.58GQPSAADD1170 pKa = 3.43MDD1172 pKa = 4.56DD1173 pKa = 3.68MNGLDD1178 pKa = 4.88DD1179 pKa = 4.15EE1180 pKa = 5.47DD1181 pKa = 4.2GVVFNNLLIAGSSGSIDD1198 pKa = 3.53VMAGSAGGQLDD1209 pKa = 3.09AWIDD1213 pKa = 3.63YY1214 pKa = 11.13GNDD1217 pKa = 2.9FSFVGDD1223 pKa = 4.21HH1224 pKa = 6.01LWGGTSHH1231 pKa = 7.64PLLPGLNAGLTFTIPAPPLAIGPVFARR1258 pKa = 11.84FRR1260 pKa = 11.84ISSGGGLAPTGLATDD1275 pKa = 4.89GEE1277 pKa = 4.67VEE1279 pKa = 4.43DD1280 pKa = 4.43YY1281 pKa = 10.79LVEE1284 pKa = 4.45LFQPAPTVPLKK1295 pKa = 9.77ITNLTFNASCTTSTVEE1311 pKa = 3.62WTVQSGITYY1320 pKa = 9.69EE1321 pKa = 4.62LEE1323 pKa = 3.95STTNLVTGPWMYY1335 pKa = 11.09VPGNPVIGPANSLTDD1350 pKa = 3.39NMSAEE1355 pKa = 3.97TSKK1358 pKa = 10.63FYY1360 pKa = 10.52RR1361 pKa = 11.84VRR1363 pKa = 11.84APYY1366 pKa = 9.59TPP1368 pKa = 3.8
MM1 pKa = 7.29KK2 pKa = 10.27AKK4 pKa = 10.36AAIAGTLLALILAGSAVAQEE24 pKa = 4.37EE25 pKa = 4.52FDD27 pKa = 5.24FGDD30 pKa = 3.97APDD33 pKa = 4.6PGFPTLLANNGARR46 pKa = 11.84HH47 pKa = 5.55LVSIGSQHH55 pKa = 6.3IFLGLAPVDD64 pKa = 4.2TEE66 pKa = 4.75PDD68 pKa = 3.66GQPHH72 pKa = 6.49INAIGDD78 pKa = 4.12DD79 pKa = 3.89NDD81 pKa = 3.46IAYY84 pKa = 7.78PPPFDD89 pKa = 6.06DD90 pKa = 4.31EE91 pKa = 6.47DD92 pKa = 4.13GVSWDD97 pKa = 3.64TPLIPGQVAQITVTASVNGDD117 pKa = 3.04LDD119 pKa = 3.31AWIDD123 pKa = 3.56FSGNNDD129 pKa = 2.69WTTVGDD135 pKa = 4.44QIFTTQPLIAGANTLNFNVPVWASPGYY162 pKa = 7.95TYY164 pKa = 11.55ARR166 pKa = 11.84FRR168 pKa = 11.84FNASGVALQHH178 pKa = 6.2VGPAPDD184 pKa = 4.91GEE186 pKa = 4.42VEE188 pKa = 4.98DD189 pKa = 4.56YY190 pKa = 10.73FIQITQEE197 pKa = 4.14LEE199 pKa = 4.3DD200 pKa = 4.67FYY202 pKa = 11.62DD203 pKa = 3.52WGDD206 pKa = 3.92APDD209 pKa = 3.95SASSPKK215 pKa = 10.43YY216 pKa = 8.36PTLAANNGANHH227 pKa = 7.84LINASVYY234 pKa = 9.96LGAVVDD240 pKa = 4.53IDD242 pKa = 6.08LDD244 pKa = 4.07GQPHH248 pKa = 6.56AKK250 pKa = 10.52ALGDD254 pKa = 5.36DD255 pKa = 3.82INQAYY260 pKa = 8.71PGSPYY265 pKa = 10.42PPGDD269 pKa = 3.3EE270 pKa = 4.96DD271 pKa = 5.5GIAFTSLLVQNQTATLTATASTAGYY296 pKa = 9.56LDD298 pKa = 3.27AWIDD302 pKa = 3.79YY303 pKa = 9.72NQDD306 pKa = 3.53GVWNAAEE313 pKa = 5.04KK314 pKa = 10.41IFSSTPLGAGANALSVTIPGGATAGKK340 pKa = 7.53TFARR344 pKa = 11.84FRR346 pKa = 11.84FSLNGGLAPNGTAPDD361 pKa = 4.29GEE363 pKa = 4.46VEE365 pKa = 4.98DD366 pKa = 4.25YY367 pKa = 10.72EE368 pKa = 4.52VWIEE372 pKa = 4.13TTEE375 pKa = 4.2EE376 pKa = 3.81DD377 pKa = 5.67HH378 pKa = 7.43KK379 pKa = 10.45MHH381 pKa = 6.91WPQYY385 pKa = 8.11PDD387 pKa = 3.08PHH389 pKa = 7.62GWDD392 pKa = 3.17VRR394 pKa = 11.84ASYY397 pKa = 11.45LDD399 pKa = 4.08GNPDD403 pKa = 3.56DD404 pKa = 4.77QFHH407 pKa = 6.84KK408 pKa = 11.09VLADD412 pKa = 4.41DD413 pKa = 5.61FLCTSNGPITKK424 pKa = 8.51ITFWGSFQYY433 pKa = 10.78NGYY436 pKa = 9.96IPTNFHH442 pKa = 6.5KK443 pKa = 10.87FNGISKK449 pKa = 8.89FHH451 pKa = 6.51FSFHH455 pKa = 6.78KK456 pKa = 10.29DD457 pKa = 3.03IPVDD461 pKa = 3.89LPEE464 pKa = 5.4IPYY467 pKa = 10.21SRR469 pKa = 11.84PVVPAEE475 pKa = 3.74WEE477 pKa = 3.82IDD479 pKa = 3.26VDD481 pKa = 4.59PFMLPAGWEE490 pKa = 3.91ITVTTEE496 pKa = 3.58EE497 pKa = 4.76PSWQGWYY504 pKa = 10.25DD505 pKa = 3.94PNTGNWNTNDD515 pKa = 3.11HH516 pKa = 5.31NHH518 pKa = 5.52YY519 pKa = 9.92FRR521 pKa = 11.84YY522 pKa = 10.42DD523 pKa = 3.2ITIPEE528 pKa = 4.28ADD530 pKa = 3.5AFVQVEE536 pKa = 4.4SNIYY540 pKa = 8.89WLDD543 pKa = 3.08ISVDD547 pKa = 3.77LEE549 pKa = 4.12GTEE552 pKa = 6.05GYY554 pKa = 9.15WGWKK558 pKa = 6.85TSRR561 pKa = 11.84SPHH564 pKa = 5.87WNDD567 pKa = 3.1DD568 pKa = 3.97AVWADD573 pKa = 3.86LPVEE577 pKa = 4.59FHH579 pKa = 5.49QQWGEE584 pKa = 4.11LIDD587 pKa = 4.82PIYY590 pKa = 11.19SNSLDD595 pKa = 3.52LAFIINDD602 pKa = 3.34EE603 pKa = 4.31VEE605 pKa = 4.15PEE607 pKa = 3.89EE608 pKa = 4.36TYY610 pKa = 11.3DD611 pKa = 3.56WGDD614 pKa = 4.23APDD617 pKa = 3.7QPYY620 pKa = 7.72PTLAASSGANHH631 pKa = 7.34LIVPGVFMGTLIDD644 pKa = 5.19AEE646 pKa = 4.44TDD648 pKa = 3.53GQPNATATGDD658 pKa = 3.96DD659 pKa = 4.0LAGPAADD666 pKa = 5.3DD667 pKa = 3.55EE668 pKa = 5.24DD669 pKa = 4.24GVVFNTPLFPGQPALIDD686 pKa = 3.6VTVSQAGWLDD696 pKa = 3.14VWLDD700 pKa = 3.34ADD702 pKa = 5.57NNGDD706 pKa = 3.36WLGPNDD712 pKa = 4.57LVYY715 pKa = 10.58SAYY718 pKa = 10.65VSAGLNPLSINIPLSVPPGQNFMRR742 pKa = 11.84FRR744 pKa = 11.84YY745 pKa = 7.45NTNGPLPPTGGAQNGEE761 pKa = 4.18VEE763 pKa = 4.7DD764 pKa = 4.34YY765 pKa = 10.92EE766 pKa = 4.6VDD768 pKa = 3.44IEE770 pKa = 5.4LEE772 pKa = 4.2PEE774 pKa = 4.31LDD776 pKa = 4.62FGDD779 pKa = 4.79APDD782 pKa = 4.55PGYY785 pKa = 8.02PTWLAMNGARR795 pKa = 11.84HH796 pKa = 6.63IIYY799 pKa = 9.96GGPAIALRR807 pKa = 11.84MGAKK811 pKa = 9.66VDD813 pKa = 4.28ADD815 pKa = 3.75PDD817 pKa = 4.11GQPTAASDD825 pKa = 4.03GDD827 pKa = 4.04DD828 pKa = 3.61TDD830 pKa = 5.04GSDD833 pKa = 3.88DD834 pKa = 3.74EE835 pKa = 4.4EE836 pKa = 5.87SIVFTTPIIPGTWAKK851 pKa = 10.01MDD853 pKa = 3.55LTVGVGIPASGYY865 pKa = 10.55AWLQGWADD873 pKa = 3.96FNNDD877 pKa = 4.03GDD879 pKa = 3.91WLDD882 pKa = 3.73AGEE885 pKa = 4.76QIFADD890 pKa = 3.71EE891 pKa = 4.72LVTFGASGSGVNNLSFPVPTTALPGALHH919 pKa = 6.25LRR921 pKa = 11.84LRR923 pKa = 11.84LSTYY927 pKa = 10.2KK928 pKa = 10.93GIGITGLAYY937 pKa = 10.4DD938 pKa = 4.95GEE940 pKa = 4.58VEE942 pKa = 4.44DD943 pKa = 4.36YY944 pKa = 10.34LVHH947 pKa = 7.78VEE949 pKa = 4.33EE950 pKa = 5.41PMDD953 pKa = 4.51IDD955 pKa = 3.84WGDD958 pKa = 4.14AFDD961 pKa = 3.89NWITPVYY968 pKa = 8.71PTILLNNGAHH978 pKa = 5.78HH979 pKa = 6.81VIDD982 pKa = 4.33GVHH985 pKa = 6.16FLGGQIDD992 pKa = 4.16AEE994 pKa = 4.34ADD996 pKa = 3.67GQPAIAVDD1004 pKa = 3.84GDD1006 pKa = 4.73DD1007 pKa = 4.22IVGPSPDD1014 pKa = 4.55DD1015 pKa = 3.44EE1016 pKa = 6.74DD1017 pKa = 5.06GVFFTTMIFAGGYY1030 pKa = 9.45VGVDD1034 pKa = 2.73VDD1036 pKa = 3.7ASTNGVLDD1044 pKa = 3.27AWVDD1048 pKa = 4.04FNNDD1052 pKa = 3.25GDD1054 pKa = 4.02WDD1056 pKa = 4.19DD1057 pKa = 5.15LNEE1060 pKa = 4.8QIFSGQNLHH1069 pKa = 6.97PGINSLGFTVPASAATVYY1087 pKa = 10.48PITSRR1092 pKa = 11.84FRR1094 pKa = 11.84FTSGGISTYY1103 pKa = 10.37TGLAMDD1109 pKa = 5.4GEE1111 pKa = 4.87VEE1113 pKa = 4.35DD1114 pKa = 4.59HH1115 pKa = 7.05ALDD1118 pKa = 3.57IQAVDD1123 pKa = 3.5TNGLDD1128 pKa = 4.33FGDD1131 pKa = 3.96ADD1133 pKa = 3.75IFPEE1137 pKa = 4.38TVLPNGARR1145 pKa = 11.84HH1146 pKa = 6.55AIVPGIVLGSAIDD1159 pKa = 3.81WEE1161 pKa = 4.67PDD1163 pKa = 3.58GQPSAADD1170 pKa = 3.43MDD1172 pKa = 4.56DD1173 pKa = 3.68MNGLDD1178 pKa = 4.88DD1179 pKa = 4.15EE1180 pKa = 5.47DD1181 pKa = 4.2GVVFNNLLIAGSSGSIDD1198 pKa = 3.53VMAGSAGGQLDD1209 pKa = 3.09AWIDD1213 pKa = 3.63YY1214 pKa = 11.13GNDD1217 pKa = 2.9FSFVGDD1223 pKa = 4.21HH1224 pKa = 6.01LWGGTSHH1231 pKa = 7.64PLLPGLNAGLTFTIPAPPLAIGPVFARR1258 pKa = 11.84FRR1260 pKa = 11.84ISSGGGLAPTGLATDD1275 pKa = 4.89GEE1277 pKa = 4.67VEE1279 pKa = 4.43DD1280 pKa = 4.43YY1281 pKa = 10.79LVEE1284 pKa = 4.45LFQPAPTVPLKK1295 pKa = 9.77ITNLTFNASCTTSTVEE1311 pKa = 3.62WTVQSGITYY1320 pKa = 9.69EE1321 pKa = 4.62LEE1323 pKa = 3.95STTNLVTGPWMYY1335 pKa = 11.09VPGNPVIGPANSLTDD1350 pKa = 3.39NMSAEE1355 pKa = 3.97TSKK1358 pKa = 10.63FYY1360 pKa = 10.52RR1361 pKa = 11.84VRR1363 pKa = 11.84APYY1366 pKa = 9.59TPP1368 pKa = 3.8
Molecular weight: 146.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6C2U951|A0A6C2U951_9BACT Uncharacterized protein OS=Pontiella desulfatans OX=2750659 GN=PDESU_04503 PE=4 SV=1
MM1 pKa = 6.56TALRR5 pKa = 11.84EE6 pKa = 4.05RR7 pKa = 11.84TIQTLRR13 pKa = 11.84LGHH16 pKa = 5.56YY17 pKa = 8.12SEE19 pKa = 5.49RR20 pKa = 11.84AIKK23 pKa = 9.6TYY25 pKa = 10.73INGLARR31 pKa = 11.84LANTTATIARR41 pKa = 11.84NLPPSASAAAPPEE54 pKa = 4.22PTGPAPWKK62 pKa = 10.3EE63 pKa = 3.7MLIISWRR70 pKa = 11.84RR71 pKa = 11.84GLALHH76 pKa = 7.12TIPIDD81 pKa = 3.84RR82 pKa = 11.84PDD84 pKa = 3.43VPGWLSPTSRR94 pKa = 11.84WHH96 pKa = 6.7LLGALISSCSLTCFSRR112 pKa = 11.84EE113 pKa = 4.13DD114 pKa = 3.24VWGSRR119 pKa = 11.84RR120 pKa = 11.84DD121 pKa = 3.28KK122 pKa = 11.01ILIVGIAA129 pKa = 3.28
MM1 pKa = 6.56TALRR5 pKa = 11.84EE6 pKa = 4.05RR7 pKa = 11.84TIQTLRR13 pKa = 11.84LGHH16 pKa = 5.56YY17 pKa = 8.12SEE19 pKa = 5.49RR20 pKa = 11.84AIKK23 pKa = 9.6TYY25 pKa = 10.73INGLARR31 pKa = 11.84LANTTATIARR41 pKa = 11.84NLPPSASAAAPPEE54 pKa = 4.22PTGPAPWKK62 pKa = 10.3EE63 pKa = 3.7MLIISWRR70 pKa = 11.84RR71 pKa = 11.84GLALHH76 pKa = 7.12TIPIDD81 pKa = 3.84RR82 pKa = 11.84PDD84 pKa = 3.43VPGWLSPTSRR94 pKa = 11.84WHH96 pKa = 6.7LLGALISSCSLTCFSRR112 pKa = 11.84EE113 pKa = 4.13DD114 pKa = 3.24VWGSRR119 pKa = 11.84RR120 pKa = 11.84DD121 pKa = 3.28KK122 pKa = 11.01ILIVGIAA129 pKa = 3.28
Molecular weight: 14.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2580498 |
29 |
6456 |
404.8 |
44.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.961 ± 0.028 | 1.173 ± 0.011 |
6.118 ± 0.023 | 6.394 ± 0.032 |
4.116 ± 0.017 | 8.512 ± 0.039 |
2.236 ± 0.015 | 5.257 ± 0.02 |
4.869 ± 0.039 | 8.948 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.537 ± 0.014 | 4.163 ± 0.025 |
4.76 ± 0.022 | 3.222 ± 0.015 |
5.156 ± 0.031 | 6.02 ± 0.034 |
5.69 ± 0.032 | 6.844 ± 0.024 |
1.735 ± 0.013 | 3.29 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
