
Acetobacterium bakii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Acetobacterium
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
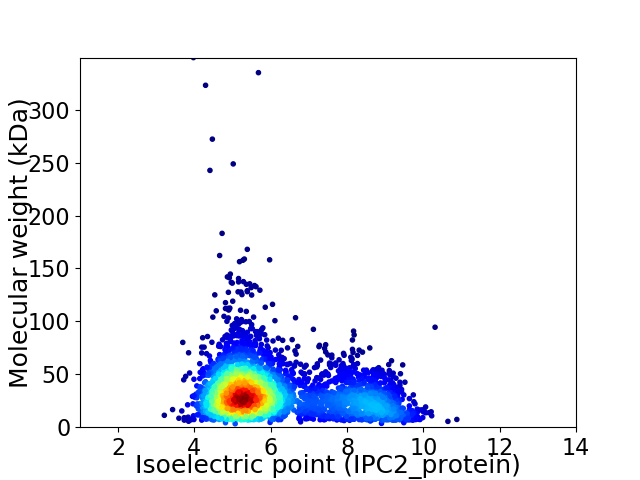
Virtual 2D-PAGE plot for 3495 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
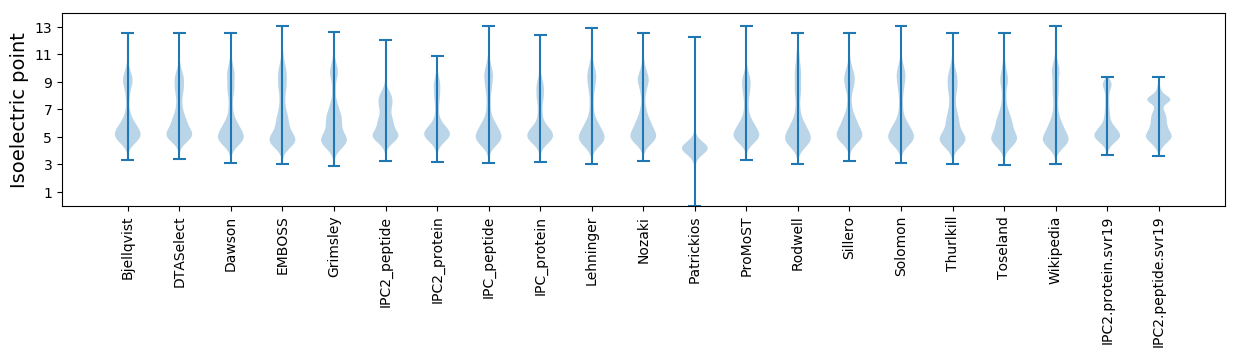
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L6U3A1|A0A0L6U3A1_9FIRM Uroporphyrinogen decarboxylase OS=Acetobacterium bakii OX=52689 GN=AKG39_07750 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.44KK3 pKa = 9.3MSKK6 pKa = 10.14FMSLLMVLIMLSMLVPVSALAADD29 pKa = 3.95NEE31 pKa = 4.33EE32 pKa = 4.33FAATEE37 pKa = 4.01DD38 pKa = 3.64NAIKK42 pKa = 10.28IVLVTKK48 pKa = 10.15EE49 pKa = 3.88AAAEE53 pKa = 3.87DD54 pKa = 4.95AIVTDD59 pKa = 4.16DD60 pKa = 3.82PVIVEE65 pKa = 4.47EE66 pKa = 4.55PLVTDD71 pKa = 4.78EE72 pKa = 4.44IVVPAEE78 pKa = 4.21SSSDD82 pKa = 3.34TAVVPEE88 pKa = 4.34QPATTEE94 pKa = 3.97INTAAVPDD102 pKa = 4.14EE103 pKa = 4.81SIVTQMASYY112 pKa = 10.28LYY114 pKa = 10.99LMQNDD119 pKa = 3.36VMIDD123 pKa = 3.44VIAANTGADD132 pKa = 3.63YY133 pKa = 10.99SYY135 pKa = 11.49DD136 pKa = 3.35AAANQLTLNNFVGEE150 pKa = 4.17NMFVLSPADD159 pKa = 3.64TFKK162 pKa = 11.5LLLLGTNILRR172 pKa = 11.84TDD174 pKa = 2.74SGFAVNVYY182 pKa = 11.18GNMNSSGTGTLIAEE196 pKa = 4.73GLATEE201 pKa = 4.21NLMGGGINVIGNLVIDD217 pKa = 3.89SGSYY221 pKa = 10.63GIVAAGFEE229 pKa = 4.39NSSSAVGILTGYY241 pKa = 10.14NDD243 pKa = 6.12DD244 pKa = 5.13IITPGNLTINGGTIDD259 pKa = 3.27IAAYY263 pKa = 10.45NSDD266 pKa = 2.91IGGAFGLFAFGDD278 pKa = 4.27LIVNGGNIDD287 pKa = 3.36IFTNSPTSYY296 pKa = 10.64SRR298 pKa = 11.84GIGAYY303 pKa = 9.84DD304 pKa = 3.09KK305 pKa = 11.41LVFNGGNTTVDD316 pKa = 3.98SITDD320 pKa = 3.35TGFAEE325 pKa = 4.8ALYY328 pKa = 10.2SYY330 pKa = 9.85NTITINNGILDD341 pKa = 5.26LYY343 pKa = 10.68TSGASANALVAEE355 pKa = 4.49GGIYY359 pKa = 10.15INPSYY364 pKa = 11.56GDD366 pKa = 3.15VDD368 pKa = 4.98LNASSLYY375 pKa = 10.68LEE377 pKa = 4.52PLVTKK382 pKa = 10.19DD383 pKa = 3.7FKK385 pKa = 10.96QPHH388 pKa = 5.87YY389 pKa = 10.79SSSNPKK395 pKa = 9.47TGVDD399 pKa = 3.7TTAMDD404 pKa = 4.17ALVAGIMIFTLIGFSFAASKK424 pKa = 10.62RR425 pKa = 11.84GAKK428 pKa = 9.48AA429 pKa = 2.83
MM1 pKa = 7.45KK2 pKa = 10.44KK3 pKa = 9.3MSKK6 pKa = 10.14FMSLLMVLIMLSMLVPVSALAADD29 pKa = 3.95NEE31 pKa = 4.33EE32 pKa = 4.33FAATEE37 pKa = 4.01DD38 pKa = 3.64NAIKK42 pKa = 10.28IVLVTKK48 pKa = 10.15EE49 pKa = 3.88AAAEE53 pKa = 3.87DD54 pKa = 4.95AIVTDD59 pKa = 4.16DD60 pKa = 3.82PVIVEE65 pKa = 4.47EE66 pKa = 4.55PLVTDD71 pKa = 4.78EE72 pKa = 4.44IVVPAEE78 pKa = 4.21SSSDD82 pKa = 3.34TAVVPEE88 pKa = 4.34QPATTEE94 pKa = 3.97INTAAVPDD102 pKa = 4.14EE103 pKa = 4.81SIVTQMASYY112 pKa = 10.28LYY114 pKa = 10.99LMQNDD119 pKa = 3.36VMIDD123 pKa = 3.44VIAANTGADD132 pKa = 3.63YY133 pKa = 10.99SYY135 pKa = 11.49DD136 pKa = 3.35AAANQLTLNNFVGEE150 pKa = 4.17NMFVLSPADD159 pKa = 3.64TFKK162 pKa = 11.5LLLLGTNILRR172 pKa = 11.84TDD174 pKa = 2.74SGFAVNVYY182 pKa = 11.18GNMNSSGTGTLIAEE196 pKa = 4.73GLATEE201 pKa = 4.21NLMGGGINVIGNLVIDD217 pKa = 3.89SGSYY221 pKa = 10.63GIVAAGFEE229 pKa = 4.39NSSSAVGILTGYY241 pKa = 10.14NDD243 pKa = 6.12DD244 pKa = 5.13IITPGNLTINGGTIDD259 pKa = 3.27IAAYY263 pKa = 10.45NSDD266 pKa = 2.91IGGAFGLFAFGDD278 pKa = 4.27LIVNGGNIDD287 pKa = 3.36IFTNSPTSYY296 pKa = 10.64SRR298 pKa = 11.84GIGAYY303 pKa = 9.84DD304 pKa = 3.09KK305 pKa = 11.41LVFNGGNTTVDD316 pKa = 3.98SITDD320 pKa = 3.35TGFAEE325 pKa = 4.8ALYY328 pKa = 10.2SYY330 pKa = 9.85NTITINNGILDD341 pKa = 5.26LYY343 pKa = 10.68TSGASANALVAEE355 pKa = 4.49GGIYY359 pKa = 10.15INPSYY364 pKa = 11.56GDD366 pKa = 3.15VDD368 pKa = 4.98LNASSLYY375 pKa = 10.68LEE377 pKa = 4.52PLVTKK382 pKa = 10.19DD383 pKa = 3.7FKK385 pKa = 10.96QPHH388 pKa = 5.87YY389 pKa = 10.79SSSNPKK395 pKa = 9.47TGVDD399 pKa = 3.7TTAMDD404 pKa = 4.17ALVAGIMIFTLIGFSFAASKK424 pKa = 10.62RR425 pKa = 11.84GAKK428 pKa = 9.48AA429 pKa = 2.83
Molecular weight: 44.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L6TXN6|A0A0L6TXN6_9FIRM Dihydrolipoyl dehydrogenase OS=Acetobacterium bakii OX=52689 GN=AKG39_14345 PE=3 SV=1
MM1 pKa = 7.87PKK3 pKa = 9.67MKK5 pKa = 8.76THH7 pKa = 6.92RR8 pKa = 11.84GAAKK12 pKa = 9.8RR13 pKa = 11.84FKK15 pKa = 9.92IKK17 pKa = 10.45KK18 pKa = 8.69SGAIKK23 pKa = 10.36RR24 pKa = 11.84FKK26 pKa = 10.49AGKK29 pKa = 8.89SHH31 pKa = 7.05ILNKK35 pKa = 9.96KK36 pKa = 6.79SRR38 pKa = 11.84KK39 pKa = 8.82RR40 pKa = 11.84KK41 pKa = 8.8RR42 pKa = 11.84NLRR45 pKa = 11.84KK46 pKa = 8.94ATGLAKK52 pKa = 10.46GDD54 pKa = 3.57AKK56 pKa = 10.48VVRR59 pKa = 11.84KK60 pKa = 8.96MISRR64 pKa = 4.2
MM1 pKa = 7.87PKK3 pKa = 9.67MKK5 pKa = 8.76THH7 pKa = 6.92RR8 pKa = 11.84GAAKK12 pKa = 9.8RR13 pKa = 11.84FKK15 pKa = 9.92IKK17 pKa = 10.45KK18 pKa = 8.69SGAIKK23 pKa = 10.36RR24 pKa = 11.84FKK26 pKa = 10.49AGKK29 pKa = 8.89SHH31 pKa = 7.05ILNKK35 pKa = 9.96KK36 pKa = 6.79SRR38 pKa = 11.84KK39 pKa = 8.82RR40 pKa = 11.84KK41 pKa = 8.8RR42 pKa = 11.84NLRR45 pKa = 11.84KK46 pKa = 8.94ATGLAKK52 pKa = 10.46GDD54 pKa = 3.57AKK56 pKa = 10.48VVRR59 pKa = 11.84KK60 pKa = 8.96MISRR64 pKa = 4.2
Molecular weight: 7.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1076816 |
29 |
3271 |
308.1 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.2 ± 0.043 | 1.286 ± 0.018 |
5.54 ± 0.034 | 6.968 ± 0.048 |
4.451 ± 0.037 | 7.137 ± 0.042 |
1.688 ± 0.019 | 8.726 ± 0.041 |
7.007 ± 0.048 | 9.467 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.035 ± 0.02 | 4.862 ± 0.032 |
3.439 ± 0.025 | 3.099 ± 0.025 |
3.717 ± 0.034 | 5.789 ± 0.032 |
5.516 ± 0.042 | 6.691 ± 0.039 |
0.783 ± 0.013 | 3.599 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
