
Monilinia fructicola (Brown rot fungus) (Ciboria fructicola)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Helotiales; Sclerotiniaceae; Monilinia
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
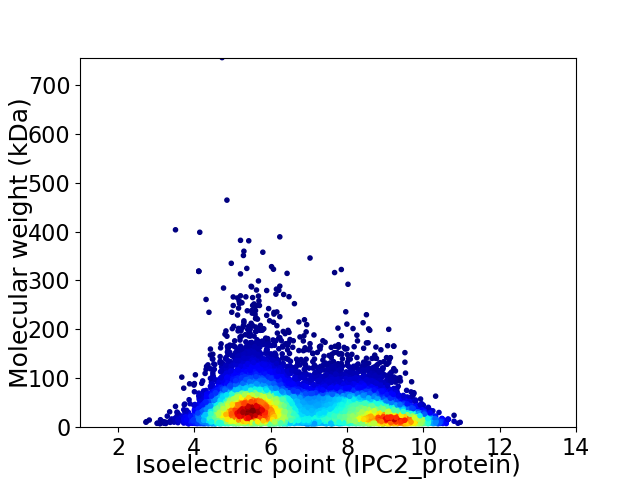
Virtual 2D-PAGE plot for 13749 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5M9JKD2|A0A5M9JKD2_MONFR Uncharacterized protein OS=Monilinia fructicola OX=38448 GN=EYC84_000790 PE=4 SV=1
MM1 pKa = 7.53SPNNPPSASSLQQEE15 pKa = 4.68TNQDD19 pKa = 3.5FPVLHH24 pKa = 5.65NRR26 pKa = 11.84KK27 pKa = 8.35MKK29 pKa = 10.05EE30 pKa = 3.57LEE32 pKa = 3.71RR33 pKa = 11.84EE34 pKa = 3.99YY35 pKa = 10.9RR36 pKa = 11.84STNRR40 pKa = 11.84TDD42 pKa = 3.47DD43 pKa = 3.46TQTNSRR49 pKa = 11.84GHH51 pKa = 5.93SAGAMAILQSFNGMKK66 pKa = 10.62GFMEE70 pKa = 4.62SYY72 pKa = 10.51GLKK75 pKa = 9.49IWLDD79 pKa = 3.42EE80 pKa = 4.3DD81 pKa = 3.79VQTARR86 pKa = 11.84QIINKK91 pKa = 7.72MLEE94 pKa = 3.67YY95 pKa = 10.62DD96 pKa = 3.81EE97 pKa = 4.35QHH99 pKa = 6.72GDD101 pKa = 3.39FGLLDD106 pKa = 3.93EE107 pKa = 5.18EE108 pKa = 4.84SADD111 pKa = 3.66GVISGRR117 pKa = 11.84EE118 pKa = 3.96FEE120 pKa = 6.02DD121 pKa = 3.46EE122 pKa = 4.14MEE124 pKa = 4.31DD125 pKa = 3.22TSRR128 pKa = 11.84IEE130 pKa = 3.84NTYY133 pKa = 11.16QNADD137 pKa = 3.66EE138 pKa = 4.45NNEE141 pKa = 3.83GSGMGNLDD149 pKa = 3.67LDD151 pKa = 3.82TRR153 pKa = 11.84QWEE156 pKa = 4.44VSHH159 pKa = 6.4GEE161 pKa = 4.27RR162 pKa = 11.84YY163 pKa = 9.49CVPDD167 pKa = 3.82EE168 pKa = 5.41LEE170 pKa = 4.67GDD172 pKa = 4.06DD173 pKa = 5.22VGGDD177 pKa = 3.99CEE179 pKa = 4.76GSCNRR184 pKa = 11.84EE185 pKa = 3.65EE186 pKa = 4.35NFDD189 pKa = 3.49VSIVDD194 pKa = 4.01RR195 pKa = 11.84EE196 pKa = 4.27EE197 pKa = 4.88GCDD200 pKa = 3.41DD201 pKa = 3.92YY202 pKa = 12.03GFGGCEE208 pKa = 4.63DD209 pKa = 5.09DD210 pKa = 5.0GQGCYY215 pKa = 10.65GDD217 pKa = 5.77DD218 pKa = 3.76YY219 pKa = 11.64DD220 pKa = 6.3GNDD223 pKa = 3.93CEE225 pKa = 5.34GYY227 pKa = 10.71DD228 pKa = 3.72YY229 pKa = 11.6GRR231 pKa = 11.84DD232 pKa = 4.24DD233 pKa = 5.55DD234 pKa = 4.17EE235 pKa = 6.98CDD237 pKa = 4.01YY238 pKa = 11.99GDD240 pKa = 5.25DD241 pKa = 3.67NCDD244 pKa = 3.3YY245 pKa = 11.36DD246 pKa = 5.15SYY248 pKa = 12.13
MM1 pKa = 7.53SPNNPPSASSLQQEE15 pKa = 4.68TNQDD19 pKa = 3.5FPVLHH24 pKa = 5.65NRR26 pKa = 11.84KK27 pKa = 8.35MKK29 pKa = 10.05EE30 pKa = 3.57LEE32 pKa = 3.71RR33 pKa = 11.84EE34 pKa = 3.99YY35 pKa = 10.9RR36 pKa = 11.84STNRR40 pKa = 11.84TDD42 pKa = 3.47DD43 pKa = 3.46TQTNSRR49 pKa = 11.84GHH51 pKa = 5.93SAGAMAILQSFNGMKK66 pKa = 10.62GFMEE70 pKa = 4.62SYY72 pKa = 10.51GLKK75 pKa = 9.49IWLDD79 pKa = 3.42EE80 pKa = 4.3DD81 pKa = 3.79VQTARR86 pKa = 11.84QIINKK91 pKa = 7.72MLEE94 pKa = 3.67YY95 pKa = 10.62DD96 pKa = 3.81EE97 pKa = 4.35QHH99 pKa = 6.72GDD101 pKa = 3.39FGLLDD106 pKa = 3.93EE107 pKa = 5.18EE108 pKa = 4.84SADD111 pKa = 3.66GVISGRR117 pKa = 11.84EE118 pKa = 3.96FEE120 pKa = 6.02DD121 pKa = 3.46EE122 pKa = 4.14MEE124 pKa = 4.31DD125 pKa = 3.22TSRR128 pKa = 11.84IEE130 pKa = 3.84NTYY133 pKa = 11.16QNADD137 pKa = 3.66EE138 pKa = 4.45NNEE141 pKa = 3.83GSGMGNLDD149 pKa = 3.67LDD151 pKa = 3.82TRR153 pKa = 11.84QWEE156 pKa = 4.44VSHH159 pKa = 6.4GEE161 pKa = 4.27RR162 pKa = 11.84YY163 pKa = 9.49CVPDD167 pKa = 3.82EE168 pKa = 5.41LEE170 pKa = 4.67GDD172 pKa = 4.06DD173 pKa = 5.22VGGDD177 pKa = 3.99CEE179 pKa = 4.76GSCNRR184 pKa = 11.84EE185 pKa = 3.65EE186 pKa = 4.35NFDD189 pKa = 3.49VSIVDD194 pKa = 4.01RR195 pKa = 11.84EE196 pKa = 4.27EE197 pKa = 4.88GCDD200 pKa = 3.41DD201 pKa = 3.92YY202 pKa = 12.03GFGGCEE208 pKa = 4.63DD209 pKa = 5.09DD210 pKa = 5.0GQGCYY215 pKa = 10.65GDD217 pKa = 5.77DD218 pKa = 3.76YY219 pKa = 11.64DD220 pKa = 6.3GNDD223 pKa = 3.93CEE225 pKa = 5.34GYY227 pKa = 10.71DD228 pKa = 3.72YY229 pKa = 11.6GRR231 pKa = 11.84DD232 pKa = 4.24DD233 pKa = 5.55DD234 pKa = 4.17EE235 pKa = 6.98CDD237 pKa = 4.01YY238 pKa = 11.99GDD240 pKa = 5.25DD241 pKa = 3.67NCDD244 pKa = 3.3YY245 pKa = 11.36DD246 pKa = 5.15SYY248 pKa = 12.13
Molecular weight: 28.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5M9K7V6|A0A5M9K7V6_MONFR Long-chain-alcohol oxidase OS=Monilinia fructicola OX=38448 GN=EYC84_006993 PE=3 SV=1
MM1 pKa = 7.56TISPSQAPTKK11 pKa = 10.15IDD13 pKa = 3.81TIPISPYY20 pKa = 9.82HH21 pKa = 6.51ALDD24 pKa = 4.22LSFLCASPSPGNSPSHH40 pKa = 6.47AAQIGKK46 pKa = 8.74PRR48 pKa = 11.84ASHH51 pKa = 6.76LSVPPPRR58 pKa = 11.84MDD60 pKa = 5.08LKK62 pKa = 9.95PTPHH66 pKa = 7.14CCTAPYY72 pKa = 10.08HH73 pKa = 6.46IFPTMRR79 pKa = 11.84LYY81 pKa = 9.54RR82 pKa = 11.84THH84 pKa = 6.64HH85 pKa = 6.49HH86 pKa = 6.97PPDD89 pKa = 3.6THH91 pKa = 6.73HH92 pKa = 7.13KK93 pKa = 10.04LHH95 pKa = 6.55TALASPTQNPNPPHH109 pKa = 7.56RR110 pKa = 11.84IRR112 pKa = 11.84IPIPIPKK119 pKa = 8.98PSHH122 pKa = 5.21TRR124 pKa = 11.84SPAKK128 pKa = 10.37KK129 pKa = 9.15EE130 pKa = 3.91AVQALPPPPSRR141 pKa = 11.84RR142 pKa = 11.84NNTNPRR148 pKa = 11.84HH149 pKa = 6.65RR150 pKa = 11.84IAQTPKK156 pKa = 10.27DD157 pKa = 3.74AKK159 pKa = 9.43KK160 pKa = 10.37CPSIIPAMRR169 pKa = 11.84RR170 pKa = 11.84HH171 pKa = 6.45PNPDD175 pKa = 2.92SCFMARR181 pKa = 11.84HH182 pKa = 5.8LPFLRR187 pKa = 11.84CMFHH191 pKa = 5.95GQRR194 pKa = 11.84GSICTSIPTLYY205 pKa = 9.54TLHH208 pKa = 6.0ITHH211 pKa = 6.56YY212 pKa = 9.4PFQPPMQHH220 pKa = 4.51RR221 pKa = 11.84HH222 pKa = 3.64MAIFRR227 pKa = 11.84VNHH230 pKa = 5.49ARR232 pKa = 11.84VNVTLPSFPPPSFPPPFLPRR252 pKa = 11.84HH253 pKa = 6.19PPPQQDD259 pKa = 2.94IALCAASPEE268 pKa = 4.23PKK270 pKa = 9.57HH271 pKa = 6.34PKK273 pKa = 9.54IIHH276 pKa = 6.28PCNDD280 pKa = 3.1TRR282 pKa = 11.84CDD284 pKa = 3.69AMGSDD289 pKa = 3.44ATRR292 pKa = 11.84RR293 pKa = 11.84DD294 pKa = 3.68TRR296 pKa = 11.84AHH298 pKa = 5.55PQKK301 pKa = 10.69SEE303 pKa = 3.78PRR305 pKa = 11.84TNSCPSVPLSPLPFLHH321 pKa = 7.16ASCFTCTSPLRR332 pKa = 11.84QPSPPRR338 pKa = 11.84RR339 pKa = 11.84TCKK342 pKa = 9.96PQAKK346 pKa = 10.01ASTIPLRR353 pKa = 11.84FHH355 pKa = 6.72RR356 pKa = 11.84VSRR359 pKa = 11.84DD360 pKa = 3.48PSVHH364 pKa = 6.18LANLTGTTRR373 pKa = 11.84PTSSNGSRR381 pKa = 11.84APDD384 pKa = 3.04RR385 pKa = 11.84MHH387 pKa = 7.1ALFSFIYY394 pKa = 9.3EE395 pKa = 4.18FYY397 pKa = 10.62ISIHH401 pKa = 6.25ASSAVGAIVQLLGCLNPNPRR421 pKa = 11.84PRR423 pKa = 11.84LNSNHH428 pKa = 6.01
MM1 pKa = 7.56TISPSQAPTKK11 pKa = 10.15IDD13 pKa = 3.81TIPISPYY20 pKa = 9.82HH21 pKa = 6.51ALDD24 pKa = 4.22LSFLCASPSPGNSPSHH40 pKa = 6.47AAQIGKK46 pKa = 8.74PRR48 pKa = 11.84ASHH51 pKa = 6.76LSVPPPRR58 pKa = 11.84MDD60 pKa = 5.08LKK62 pKa = 9.95PTPHH66 pKa = 7.14CCTAPYY72 pKa = 10.08HH73 pKa = 6.46IFPTMRR79 pKa = 11.84LYY81 pKa = 9.54RR82 pKa = 11.84THH84 pKa = 6.64HH85 pKa = 6.49HH86 pKa = 6.97PPDD89 pKa = 3.6THH91 pKa = 6.73HH92 pKa = 7.13KK93 pKa = 10.04LHH95 pKa = 6.55TALASPTQNPNPPHH109 pKa = 7.56RR110 pKa = 11.84IRR112 pKa = 11.84IPIPIPKK119 pKa = 8.98PSHH122 pKa = 5.21TRR124 pKa = 11.84SPAKK128 pKa = 10.37KK129 pKa = 9.15EE130 pKa = 3.91AVQALPPPPSRR141 pKa = 11.84RR142 pKa = 11.84NNTNPRR148 pKa = 11.84HH149 pKa = 6.65RR150 pKa = 11.84IAQTPKK156 pKa = 10.27DD157 pKa = 3.74AKK159 pKa = 9.43KK160 pKa = 10.37CPSIIPAMRR169 pKa = 11.84RR170 pKa = 11.84HH171 pKa = 6.45PNPDD175 pKa = 2.92SCFMARR181 pKa = 11.84HH182 pKa = 5.8LPFLRR187 pKa = 11.84CMFHH191 pKa = 5.95GQRR194 pKa = 11.84GSICTSIPTLYY205 pKa = 9.54TLHH208 pKa = 6.0ITHH211 pKa = 6.56YY212 pKa = 9.4PFQPPMQHH220 pKa = 4.51RR221 pKa = 11.84HH222 pKa = 3.64MAIFRR227 pKa = 11.84VNHH230 pKa = 5.49ARR232 pKa = 11.84VNVTLPSFPPPSFPPPFLPRR252 pKa = 11.84HH253 pKa = 6.19PPPQQDD259 pKa = 2.94IALCAASPEE268 pKa = 4.23PKK270 pKa = 9.57HH271 pKa = 6.34PKK273 pKa = 9.54IIHH276 pKa = 6.28PCNDD280 pKa = 3.1TRR282 pKa = 11.84CDD284 pKa = 3.69AMGSDD289 pKa = 3.44ATRR292 pKa = 11.84RR293 pKa = 11.84DD294 pKa = 3.68TRR296 pKa = 11.84AHH298 pKa = 5.55PQKK301 pKa = 10.69SEE303 pKa = 3.78PRR305 pKa = 11.84TNSCPSVPLSPLPFLHH321 pKa = 7.16ASCFTCTSPLRR332 pKa = 11.84QPSPPRR338 pKa = 11.84RR339 pKa = 11.84TCKK342 pKa = 9.96PQAKK346 pKa = 10.01ASTIPLRR353 pKa = 11.84FHH355 pKa = 6.72RR356 pKa = 11.84VSRR359 pKa = 11.84DD360 pKa = 3.48PSVHH364 pKa = 6.18LANLTGTTRR373 pKa = 11.84PTSSNGSRR381 pKa = 11.84APDD384 pKa = 3.04RR385 pKa = 11.84MHH387 pKa = 7.1ALFSFIYY394 pKa = 9.3EE395 pKa = 4.18FYY397 pKa = 10.62ISIHH401 pKa = 6.25ASSAVGAIVQLLGCLNPNPRR421 pKa = 11.84PRR423 pKa = 11.84LNSNHH428 pKa = 6.01
Molecular weight: 47.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5524529 |
66 |
6810 |
401.8 |
44.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.694 ± 0.019 | 1.201 ± 0.008 |
5.446 ± 0.016 | 6.429 ± 0.029 |
3.708 ± 0.014 | 6.795 ± 0.025 |
2.414 ± 0.01 | 5.473 ± 0.017 |
5.387 ± 0.021 | 8.535 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.209 ± 0.008 | 4.165 ± 0.013 |
5.978 ± 0.025 | 3.905 ± 0.018 |
5.85 ± 0.02 | 8.862 ± 0.031 |
6.127 ± 0.017 | 5.67 ± 0.015 |
1.36 ± 0.007 | 2.79 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
