
Massilia timonae CCUG 45783
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Massilia; Massilia timonae
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
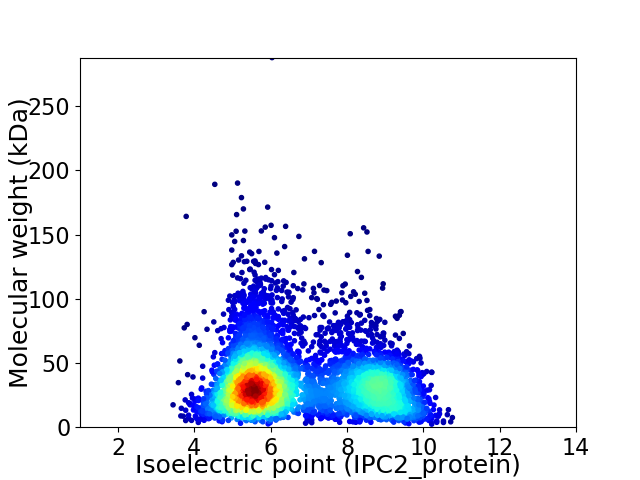
Virtual 2D-PAGE plot for 5096 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9DBE1|K9DBE1_9BURK HTH lysR-type domain-containing protein OS=Massilia timonae CCUG 45783 OX=883126 GN=HMPREF9710_02687 PE=4 SV=1
MM1 pKa = 6.79TAVAEE6 pKa = 4.31VQDD9 pKa = 4.51FDD11 pKa = 4.94TIPVPINFTDD21 pKa = 3.53SAAQKK26 pKa = 9.24VAQLIEE32 pKa = 4.37EE33 pKa = 4.51EE34 pKa = 4.63GNPDD38 pKa = 2.85LKK40 pKa = 11.13LRR42 pKa = 11.84VFVQGGGCSGFQYY55 pKa = 10.94GFTFDD60 pKa = 4.69EE61 pKa = 4.93IVNEE65 pKa = 4.96DD66 pKa = 3.85DD67 pKa = 3.19TTMEE71 pKa = 4.19KK72 pKa = 10.82NGVQLLIDD80 pKa = 3.77SMSYY84 pKa = 10.23QYY86 pKa = 11.55LVGAEE91 pKa = 4.01IDD93 pKa = 3.89YY94 pKa = 11.28KK95 pKa = 11.39DD96 pKa = 4.47DD97 pKa = 4.08LEE99 pKa = 4.56GAQFVIKK106 pKa = 10.53NPNATSTCGCGSSFSAA122 pKa = 4.79
MM1 pKa = 6.79TAVAEE6 pKa = 4.31VQDD9 pKa = 4.51FDD11 pKa = 4.94TIPVPINFTDD21 pKa = 3.53SAAQKK26 pKa = 9.24VAQLIEE32 pKa = 4.37EE33 pKa = 4.51EE34 pKa = 4.63GNPDD38 pKa = 2.85LKK40 pKa = 11.13LRR42 pKa = 11.84VFVQGGGCSGFQYY55 pKa = 10.94GFTFDD60 pKa = 4.69EE61 pKa = 4.93IVNEE65 pKa = 4.96DD66 pKa = 3.85DD67 pKa = 3.19TTMEE71 pKa = 4.19KK72 pKa = 10.82NGVQLLIDD80 pKa = 3.77SMSYY84 pKa = 10.23QYY86 pKa = 11.55LVGAEE91 pKa = 4.01IDD93 pKa = 3.89YY94 pKa = 11.28KK95 pKa = 11.39DD96 pKa = 4.47DD97 pKa = 4.08LEE99 pKa = 4.56GAQFVIKK106 pKa = 10.53NPNATSTCGCGSSFSAA122 pKa = 4.79
Molecular weight: 13.22 kDa
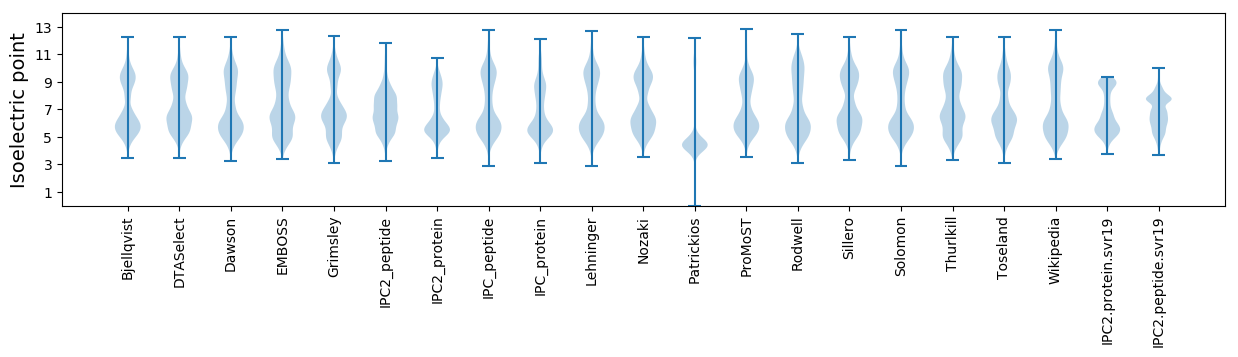
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9DBC5|K9DBC5_9BURK Protein-tyrosine-phosphatase OS=Massilia timonae CCUG 45783 OX=883126 GN=HMPREF9710_04115 PE=4 SV=1
MM1 pKa = 7.85KK2 pKa = 10.37APQFEE7 pKa = 4.56KK8 pKa = 10.99LFAQLALLNQPQHH21 pKa = 6.04RR22 pKa = 11.84QVLVALHH29 pKa = 6.6PAAGLDD35 pKa = 3.27RR36 pKa = 11.84VVALIGEE43 pKa = 4.29IRR45 pKa = 11.84SKK47 pKa = 10.94GRR49 pKa = 11.84RR50 pKa = 11.84CPDD53 pKa = 3.54CACDD57 pKa = 3.02RR58 pKa = 11.84CHH60 pKa = 7.38RR61 pKa = 11.84HH62 pKa = 5.26GQANDD67 pKa = 3.32LQRR70 pKa = 11.84YY71 pKa = 6.7RR72 pKa = 11.84CCACGRR78 pKa = 11.84TFNDD82 pKa = 3.34LTGTPLARR90 pKa = 11.84LRR92 pKa = 11.84HH93 pKa = 5.26KK94 pKa = 10.61GKK96 pKa = 9.19WLEE99 pKa = 4.03YY100 pKa = 10.25LDD102 pKa = 4.23TVLDD106 pKa = 3.81SRR108 pKa = 11.84TVRR111 pKa = 11.84SAAKK115 pKa = 10.1RR116 pKa = 11.84IGVHH120 pKa = 6.59RR121 pKa = 11.84NTTFRR126 pKa = 11.84WRR128 pKa = 11.84HH129 pKa = 4.41RR130 pKa = 11.84VLDD133 pKa = 3.8RR134 pKa = 11.84VKK136 pKa = 10.87DD137 pKa = 3.78DD138 pKa = 4.53RR139 pKa = 11.84PDD141 pKa = 3.36HH142 pKa = 6.22LVGIVEE148 pKa = 4.21ADD150 pKa = 3.3EE151 pKa = 4.41MFLLEE156 pKa = 4.14SQKK159 pKa = 10.8GSRR162 pKa = 11.84KK163 pKa = 9.53LDD165 pKa = 3.32RR166 pKa = 11.84PPRR169 pKa = 11.84KK170 pKa = 9.42RR171 pKa = 11.84GGRR174 pKa = 11.84AALRR178 pKa = 11.84GISHH182 pKa = 7.15HH183 pKa = 6.89LDD185 pKa = 4.37CILVARR191 pKa = 11.84DD192 pKa = 3.6RR193 pKa = 11.84GGQTIDD199 pKa = 3.03AVTGRR204 pKa = 11.84GALKK208 pKa = 9.66VAQLVTHH215 pKa = 7.06LLPKK219 pKa = 10.5LHH221 pKa = 6.51AQALLVTDD229 pKa = 5.2ANTAYY234 pKa = 9.78PAFARR239 pKa = 11.84AHH241 pKa = 5.58GVAHH245 pKa = 5.58QAVNLSAGEE254 pKa = 4.04RR255 pKa = 11.84VRR257 pKa = 11.84KK258 pKa = 9.79GDD260 pKa = 3.11AGAIHH265 pKa = 5.77VQNVNAYY272 pKa = 9.0HH273 pKa = 6.79RR274 pKa = 11.84RR275 pKa = 11.84FKK277 pKa = 10.56EE278 pKa = 3.84WLAHH282 pKa = 4.55FHH284 pKa = 6.16GVASRR289 pKa = 11.84WLPNYY294 pKa = 10.14LGWHH298 pKa = 5.85WALDD302 pKa = 3.7GGRR305 pKa = 11.84VNSVEE310 pKa = 3.79QLLRR314 pKa = 11.84IAFGVINSS322 pKa = 3.73
MM1 pKa = 7.85KK2 pKa = 10.37APQFEE7 pKa = 4.56KK8 pKa = 10.99LFAQLALLNQPQHH21 pKa = 6.04RR22 pKa = 11.84QVLVALHH29 pKa = 6.6PAAGLDD35 pKa = 3.27RR36 pKa = 11.84VVALIGEE43 pKa = 4.29IRR45 pKa = 11.84SKK47 pKa = 10.94GRR49 pKa = 11.84RR50 pKa = 11.84CPDD53 pKa = 3.54CACDD57 pKa = 3.02RR58 pKa = 11.84CHH60 pKa = 7.38RR61 pKa = 11.84HH62 pKa = 5.26GQANDD67 pKa = 3.32LQRR70 pKa = 11.84YY71 pKa = 6.7RR72 pKa = 11.84CCACGRR78 pKa = 11.84TFNDD82 pKa = 3.34LTGTPLARR90 pKa = 11.84LRR92 pKa = 11.84HH93 pKa = 5.26KK94 pKa = 10.61GKK96 pKa = 9.19WLEE99 pKa = 4.03YY100 pKa = 10.25LDD102 pKa = 4.23TVLDD106 pKa = 3.81SRR108 pKa = 11.84TVRR111 pKa = 11.84SAAKK115 pKa = 10.1RR116 pKa = 11.84IGVHH120 pKa = 6.59RR121 pKa = 11.84NTTFRR126 pKa = 11.84WRR128 pKa = 11.84HH129 pKa = 4.41RR130 pKa = 11.84VLDD133 pKa = 3.8RR134 pKa = 11.84VKK136 pKa = 10.87DD137 pKa = 3.78DD138 pKa = 4.53RR139 pKa = 11.84PDD141 pKa = 3.36HH142 pKa = 6.22LVGIVEE148 pKa = 4.21ADD150 pKa = 3.3EE151 pKa = 4.41MFLLEE156 pKa = 4.14SQKK159 pKa = 10.8GSRR162 pKa = 11.84KK163 pKa = 9.53LDD165 pKa = 3.32RR166 pKa = 11.84PPRR169 pKa = 11.84KK170 pKa = 9.42RR171 pKa = 11.84GGRR174 pKa = 11.84AALRR178 pKa = 11.84GISHH182 pKa = 7.15HH183 pKa = 6.89LDD185 pKa = 4.37CILVARR191 pKa = 11.84DD192 pKa = 3.6RR193 pKa = 11.84GGQTIDD199 pKa = 3.03AVTGRR204 pKa = 11.84GALKK208 pKa = 9.66VAQLVTHH215 pKa = 7.06LLPKK219 pKa = 10.5LHH221 pKa = 6.51AQALLVTDD229 pKa = 5.2ANTAYY234 pKa = 9.78PAFARR239 pKa = 11.84AHH241 pKa = 5.58GVAHH245 pKa = 5.58QAVNLSAGEE254 pKa = 4.04RR255 pKa = 11.84VRR257 pKa = 11.84KK258 pKa = 9.79GDD260 pKa = 3.11AGAIHH265 pKa = 5.77VQNVNAYY272 pKa = 9.0HH273 pKa = 6.79RR274 pKa = 11.84RR275 pKa = 11.84FKK277 pKa = 10.56EE278 pKa = 3.84WLAHH282 pKa = 4.55FHH284 pKa = 6.16GVASRR289 pKa = 11.84WLPNYY294 pKa = 10.14LGWHH298 pKa = 5.85WALDD302 pKa = 3.7GGRR305 pKa = 11.84VNSVEE310 pKa = 3.79QLLRR314 pKa = 11.84IAFGVINSS322 pKa = 3.73
Molecular weight: 36.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1765012 |
23 |
2700 |
346.4 |
37.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.736 ± 0.052 | 0.858 ± 0.011 |
5.575 ± 0.023 | 5.388 ± 0.029 |
3.516 ± 0.02 | 8.393 ± 0.028 |
2.238 ± 0.017 | 4.513 ± 0.026 |
3.305 ± 0.034 | 10.465 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.487 ± 0.015 | 2.894 ± 0.022 |
5.145 ± 0.023 | 3.75 ± 0.022 |
7.101 ± 0.035 | 5.325 ± 0.027 |
5.07 ± 0.028 | 7.418 ± 0.027 |
1.377 ± 0.013 | 2.444 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
