
Lactobacillus phage 521B
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Herelleviridae; Tybeckvirus; Lactobacillus virus 521B
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
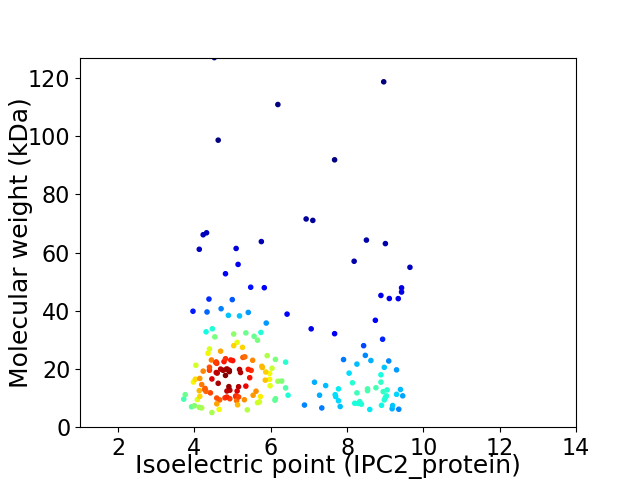
Virtual 2D-PAGE plot for 188 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y5FEG1|A0A4Y5FEG1_9CAUD Transposase OS=Lactobacillus phage 521B OX=2510942 GN=B521_0072 PE=3 SV=1
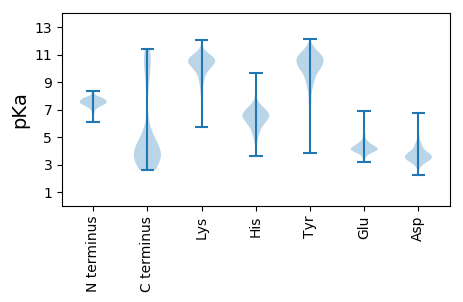
MM1 pKa = 7.46TNQSIKK7 pKa = 10.66AYY9 pKa = 9.68FYY11 pKa = 11.16NGFNEE16 pKa = 4.36PEE18 pKa = 4.82LIATDD23 pKa = 3.57IEE25 pKa = 4.32EE26 pKa = 4.36LKK28 pKa = 10.96HH29 pKa = 6.88VIIYY33 pKa = 10.4DD34 pKa = 3.6NYY36 pKa = 10.17IGDD39 pKa = 3.71VSKK42 pKa = 11.43DD43 pKa = 3.53LLDD46 pKa = 5.87DD47 pKa = 4.25INSCSDD53 pKa = 3.5LDD55 pKa = 4.01DD56 pKa = 4.84LKK58 pKa = 11.62EE59 pKa = 3.78EE60 pKa = 4.28LAYY63 pKa = 10.49FDD65 pKa = 5.38IFINPDD71 pKa = 3.04KK72 pKa = 11.05QDD74 pKa = 4.13FIEE77 pKa = 4.96ANKK80 pKa = 10.65AWFEE84 pKa = 3.96PDD86 pKa = 2.97GLEE89 pKa = 4.9NYY91 pKa = 10.11LEE93 pKa = 4.13NWW95 pKa = 3.31
MM1 pKa = 7.46TNQSIKK7 pKa = 10.66AYY9 pKa = 9.68FYY11 pKa = 11.16NGFNEE16 pKa = 4.36PEE18 pKa = 4.82LIATDD23 pKa = 3.57IEE25 pKa = 4.32EE26 pKa = 4.36LKK28 pKa = 10.96HH29 pKa = 6.88VIIYY33 pKa = 10.4DD34 pKa = 3.6NYY36 pKa = 10.17IGDD39 pKa = 3.71VSKK42 pKa = 11.43DD43 pKa = 3.53LLDD46 pKa = 5.87DD47 pKa = 4.25INSCSDD53 pKa = 3.5LDD55 pKa = 4.01DD56 pKa = 4.84LKK58 pKa = 11.62EE59 pKa = 3.78EE60 pKa = 4.28LAYY63 pKa = 10.49FDD65 pKa = 5.38IFINPDD71 pKa = 3.04KK72 pKa = 11.05QDD74 pKa = 4.13FIEE77 pKa = 4.96ANKK80 pKa = 10.65AWFEE84 pKa = 3.96PDD86 pKa = 2.97GLEE89 pKa = 4.9NYY91 pKa = 10.11LEE93 pKa = 4.13NWW95 pKa = 3.31
Molecular weight: 11.18 kDa
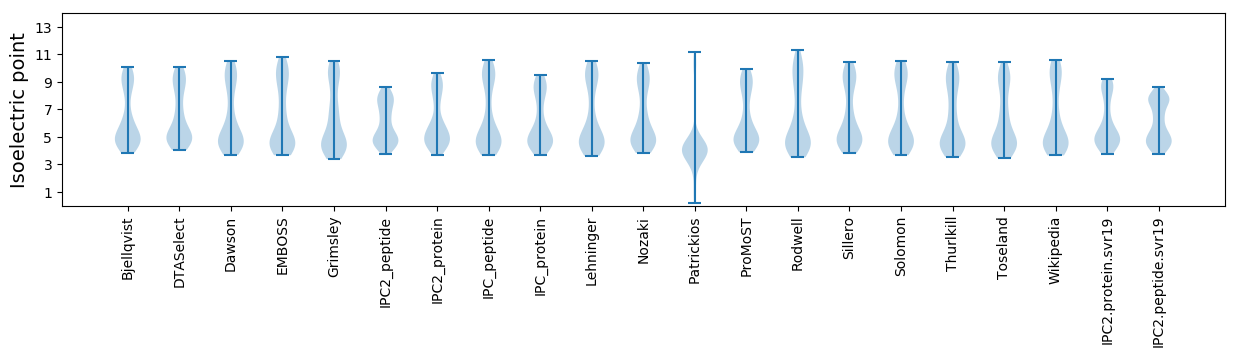
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y5FED1|A0A4Y5FED1_9CAUD Uncharacterized protein OS=Lactobacillus phage 521B OX=2510942 GN=B521_0051 PE=4 SV=1
MM1 pKa = 6.67LTSYY5 pKa = 10.46KK6 pKa = 10.45VEE8 pKa = 4.25INPTSQQAWLIDD20 pKa = 3.46RR21 pKa = 11.84HH22 pKa = 6.54ISASRR27 pKa = 11.84WAYY30 pKa = 11.43NLFLDD35 pKa = 4.63INKK38 pKa = 8.6QRR40 pKa = 11.84YY41 pKa = 7.37EE42 pKa = 4.05DD43 pKa = 4.44GYY45 pKa = 10.61HH46 pKa = 6.43YY47 pKa = 10.37MSAYY51 pKa = 9.78EE52 pKa = 3.72FSKK55 pKa = 10.21WFNHH59 pKa = 7.38DD60 pKa = 3.77YY61 pKa = 11.56LDD63 pKa = 5.43FNPDD67 pKa = 3.76DD68 pKa = 3.54NWIKK72 pKa = 10.44QLYY75 pKa = 9.48AKK77 pKa = 8.63STKK80 pKa = 9.42QAFIDD85 pKa = 4.09ADD87 pKa = 3.27NSMKK91 pKa = 10.55RR92 pKa = 11.84FFKK95 pKa = 10.47KK96 pKa = 9.5QSRR99 pKa = 11.84YY100 pKa = 7.51PHH102 pKa = 5.8FRR104 pKa = 11.84SFKK107 pKa = 8.39RR108 pKa = 11.84GKK110 pKa = 9.84GSFYY114 pKa = 10.57FVKK117 pKa = 10.61NSNKK121 pKa = 10.33DD122 pKa = 3.17IIRR125 pKa = 11.84VEE127 pKa = 3.71RR128 pKa = 11.84HH129 pKa = 5.95RR130 pKa = 11.84IKK132 pKa = 10.57LPKK135 pKa = 9.72IGWVRR140 pKa = 11.84LKK142 pKa = 10.61EE143 pKa = 3.92KK144 pKa = 10.73GYY146 pKa = 10.14IPTDD150 pKa = 3.37AEE152 pKa = 4.16HH153 pKa = 6.73FVIKK157 pKa = 10.12QGRR160 pKa = 11.84VKK162 pKa = 10.32KK163 pKa = 10.04HH164 pKa = 5.57AGRR167 pKa = 11.84YY168 pKa = 8.77YY169 pKa = 9.42LTCLVEE175 pKa = 3.87QADD178 pKa = 3.69IQFAKK183 pKa = 10.9LNGDD187 pKa = 4.18PIGIDD192 pKa = 3.26LGIKK196 pKa = 9.74EE197 pKa = 4.48FAILSNGTIFRR208 pKa = 11.84NKK210 pKa = 9.62NKK212 pKa = 9.8SSRR215 pKa = 11.84VRR217 pKa = 11.84GMRR220 pKa = 11.84KK221 pKa = 9.14RR222 pKa = 11.84LRR224 pKa = 11.84RR225 pKa = 11.84LQRR228 pKa = 11.84KK229 pKa = 6.98WSRR232 pKa = 11.84QYY234 pKa = 11.38LAFKK238 pKa = 10.39ARR240 pKa = 11.84KK241 pKa = 8.33QKK243 pKa = 10.47EE244 pKa = 4.27GSSATDD250 pKa = 3.53LNLAKK255 pKa = 9.89TKK257 pKa = 11.05LKK259 pKa = 9.07MQRR262 pKa = 11.84LYY264 pKa = 10.99QRR266 pKa = 11.84LQNVQTDD273 pKa = 3.99YY274 pKa = 10.83QKK276 pKa = 11.22KK277 pKa = 9.9IISQVVRR284 pKa = 11.84TKK286 pKa = 9.7PQWVALEE293 pKa = 4.26DD294 pKa = 3.76LNIRR298 pKa = 11.84GMAKK302 pKa = 10.04NKK304 pKa = 9.85HH305 pKa = 4.65LAKK308 pKa = 10.59AIMQQGFYY316 pKa = 10.97AFRR319 pKa = 11.84EE320 pKa = 3.99KK321 pKa = 10.83LINKK325 pKa = 7.38CQQLGIPVHH334 pKa = 6.46LINRR338 pKa = 11.84FEE340 pKa = 4.14PTSKK344 pKa = 10.43CCHH347 pKa = 5.6NCGARR352 pKa = 11.84KK353 pKa = 10.04VNLSLSDD360 pKa = 4.54RR361 pKa = 11.84IYY363 pKa = 10.32HH364 pKa = 6.69CDD366 pKa = 3.02VCGYY370 pKa = 9.37TEE372 pKa = 5.06DD373 pKa = 4.67RR374 pKa = 11.84DD375 pKa = 3.81INASLNIRR383 pKa = 11.84DD384 pKa = 3.89TEE386 pKa = 4.14NFEE389 pKa = 4.16LAYY392 pKa = 10.5
MM1 pKa = 6.67LTSYY5 pKa = 10.46KK6 pKa = 10.45VEE8 pKa = 4.25INPTSQQAWLIDD20 pKa = 3.46RR21 pKa = 11.84HH22 pKa = 6.54ISASRR27 pKa = 11.84WAYY30 pKa = 11.43NLFLDD35 pKa = 4.63INKK38 pKa = 8.6QRR40 pKa = 11.84YY41 pKa = 7.37EE42 pKa = 4.05DD43 pKa = 4.44GYY45 pKa = 10.61HH46 pKa = 6.43YY47 pKa = 10.37MSAYY51 pKa = 9.78EE52 pKa = 3.72FSKK55 pKa = 10.21WFNHH59 pKa = 7.38DD60 pKa = 3.77YY61 pKa = 11.56LDD63 pKa = 5.43FNPDD67 pKa = 3.76DD68 pKa = 3.54NWIKK72 pKa = 10.44QLYY75 pKa = 9.48AKK77 pKa = 8.63STKK80 pKa = 9.42QAFIDD85 pKa = 4.09ADD87 pKa = 3.27NSMKK91 pKa = 10.55RR92 pKa = 11.84FFKK95 pKa = 10.47KK96 pKa = 9.5QSRR99 pKa = 11.84YY100 pKa = 7.51PHH102 pKa = 5.8FRR104 pKa = 11.84SFKK107 pKa = 8.39RR108 pKa = 11.84GKK110 pKa = 9.84GSFYY114 pKa = 10.57FVKK117 pKa = 10.61NSNKK121 pKa = 10.33DD122 pKa = 3.17IIRR125 pKa = 11.84VEE127 pKa = 3.71RR128 pKa = 11.84HH129 pKa = 5.95RR130 pKa = 11.84IKK132 pKa = 10.57LPKK135 pKa = 9.72IGWVRR140 pKa = 11.84LKK142 pKa = 10.61EE143 pKa = 3.92KK144 pKa = 10.73GYY146 pKa = 10.14IPTDD150 pKa = 3.37AEE152 pKa = 4.16HH153 pKa = 6.73FVIKK157 pKa = 10.12QGRR160 pKa = 11.84VKK162 pKa = 10.32KK163 pKa = 10.04HH164 pKa = 5.57AGRR167 pKa = 11.84YY168 pKa = 8.77YY169 pKa = 9.42LTCLVEE175 pKa = 3.87QADD178 pKa = 3.69IQFAKK183 pKa = 10.9LNGDD187 pKa = 4.18PIGIDD192 pKa = 3.26LGIKK196 pKa = 9.74EE197 pKa = 4.48FAILSNGTIFRR208 pKa = 11.84NKK210 pKa = 9.62NKK212 pKa = 9.8SSRR215 pKa = 11.84VRR217 pKa = 11.84GMRR220 pKa = 11.84KK221 pKa = 9.14RR222 pKa = 11.84LRR224 pKa = 11.84RR225 pKa = 11.84LQRR228 pKa = 11.84KK229 pKa = 6.98WSRR232 pKa = 11.84QYY234 pKa = 11.38LAFKK238 pKa = 10.39ARR240 pKa = 11.84KK241 pKa = 8.33QKK243 pKa = 10.47EE244 pKa = 4.27GSSATDD250 pKa = 3.53LNLAKK255 pKa = 9.89TKK257 pKa = 11.05LKK259 pKa = 9.07MQRR262 pKa = 11.84LYY264 pKa = 10.99QRR266 pKa = 11.84LQNVQTDD273 pKa = 3.99YY274 pKa = 10.83QKK276 pKa = 11.22KK277 pKa = 9.9IISQVVRR284 pKa = 11.84TKK286 pKa = 9.7PQWVALEE293 pKa = 4.26DD294 pKa = 3.76LNIRR298 pKa = 11.84GMAKK302 pKa = 10.04NKK304 pKa = 9.85HH305 pKa = 4.65LAKK308 pKa = 10.59AIMQQGFYY316 pKa = 10.97AFRR319 pKa = 11.84EE320 pKa = 3.99KK321 pKa = 10.83LINKK325 pKa = 7.38CQQLGIPVHH334 pKa = 6.46LINRR338 pKa = 11.84FEE340 pKa = 4.14PTSKK344 pKa = 10.43CCHH347 pKa = 5.6NCGARR352 pKa = 11.84KK353 pKa = 10.04VNLSLSDD360 pKa = 4.54RR361 pKa = 11.84IYY363 pKa = 10.32HH364 pKa = 6.69CDD366 pKa = 3.02VCGYY370 pKa = 9.37TEE372 pKa = 5.06DD373 pKa = 4.67RR374 pKa = 11.84DD375 pKa = 3.81INASLNIRR383 pKa = 11.84DD384 pKa = 3.89TEE386 pKa = 4.14NFEE389 pKa = 4.16LAYY392 pKa = 10.5
Molecular weight: 46.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
40347 |
43 |
1169 |
214.6 |
24.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.838 ± 0.209 | 0.617 ± 0.059 |
7.264 ± 0.175 | 5.929 ± 0.25 |
4.008 ± 0.122 | 5.547 ± 0.235 |
1.376 ± 0.089 | 7.683 ± 0.194 |
8.685 ± 0.282 | 8.291 ± 0.187 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.077 | 7.773 ± 0.184 |
2.922 ± 0.14 | 3.222 ± 0.154 |
3.066 ± 0.149 | 8.219 ± 0.313 |
5.938 ± 0.243 | 5.805 ± 0.135 |
1.068 ± 0.073 | 5.517 ± 0.154 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
