
Alloiococcus otitis ATCC 51267
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Alloiococcus; Alloiococcus otitis
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
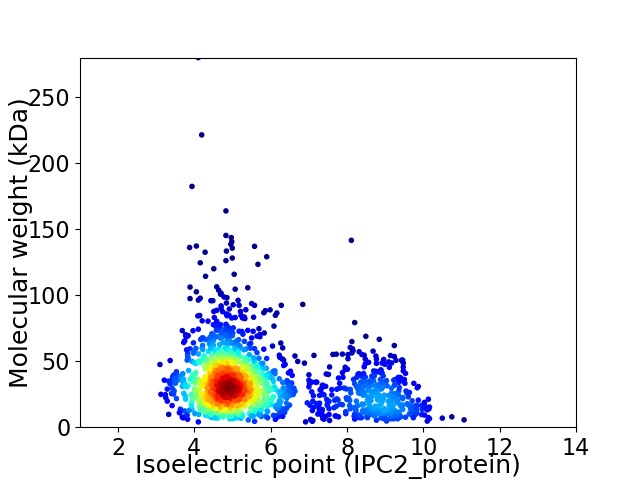
Virtual 2D-PAGE plot for 1627 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9EUT4|K9EUT4_9LACT Uncharacterized protein OS=Alloiococcus otitis ATCC 51267 OX=883081 GN=HMPREF9698_01533 PE=3 SV=1
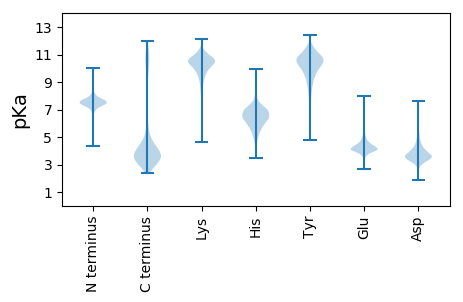
MM1 pKa = 7.34SLKK4 pKa = 10.27KK5 pKa = 10.25KK6 pKa = 10.21YY7 pKa = 10.59AILASSAALSLTLAACGGEE26 pKa = 4.39GEE28 pKa = 4.66PVDD31 pKa = 4.31PGQEE35 pKa = 4.21GEE37 pKa = 4.39SSQEE41 pKa = 3.93TTEE44 pKa = 4.24DD45 pKa = 2.99QATQEE50 pKa = 4.2RR51 pKa = 11.84SPEE54 pKa = 4.11DD55 pKa = 3.26FDD57 pKa = 3.91EE58 pKa = 4.65TVQNEE63 pKa = 4.22GEE65 pKa = 4.56AIDD68 pKa = 4.76GGIAQVGVLSDD79 pKa = 3.79TAWTGIFDD87 pKa = 3.64WQHH90 pKa = 5.73YY91 pKa = 8.15EE92 pKa = 4.0NEE94 pKa = 4.13ADD96 pKa = 3.58ASLMNYY102 pKa = 8.38TLGDD106 pKa = 4.06LLAMNEE112 pKa = 3.95NYY114 pKa = 10.37EE115 pKa = 4.18LVGGKK120 pKa = 9.85EE121 pKa = 3.71NGAAADD127 pKa = 3.67IEE129 pKa = 4.22FDD131 pKa = 3.77EE132 pKa = 6.23DD133 pKa = 3.79NDD135 pKa = 4.26TVTITVHH142 pKa = 7.51DD143 pKa = 4.04DD144 pKa = 3.96VYY146 pKa = 10.79WHH148 pKa = 7.5DD149 pKa = 4.24GEE151 pKa = 5.39QVTADD156 pKa = 4.35DD157 pKa = 3.86IVFAHH162 pKa = 7.02EE163 pKa = 4.04IVGNPDD169 pKa = 3.1YY170 pKa = 11.26TGIRR174 pKa = 11.84YY175 pKa = 9.58GDD177 pKa = 3.67NFTNVQGMEE186 pKa = 4.07EE187 pKa = 4.41YY188 pKa = 10.52KK189 pKa = 10.84AGEE192 pKa = 4.13ADD194 pKa = 3.92SISGLEE200 pKa = 4.07LSDD203 pKa = 5.85DD204 pKa = 3.69EE205 pKa = 4.59MTVTIHH211 pKa = 5.42YY212 pKa = 8.21QQVVPSMKK220 pKa = 9.55QAGRR224 pKa = 11.84GVWAYY229 pKa = 9.69AAPRR233 pKa = 11.84HH234 pKa = 5.1YY235 pKa = 11.03LEE237 pKa = 6.27DD238 pKa = 3.56IPADD242 pKa = 3.58EE243 pKa = 5.05LVSHH247 pKa = 6.65PNVRR251 pKa = 11.84EE252 pKa = 3.82NPIGFGPFKK261 pKa = 10.59VDD263 pKa = 2.93NVVPGEE269 pKa = 4.17SVEE272 pKa = 3.93YY273 pKa = 10.1SAYY276 pKa = 10.05EE277 pKa = 4.28DD278 pKa = 3.69YY279 pKa = 11.23HH280 pKa = 8.16LGEE283 pKa = 5.1PKK285 pKa = 10.43LDD287 pKa = 4.61GITLEE292 pKa = 4.13RR293 pKa = 11.84LQPAGAIEE301 pKa = 4.03ALRR304 pKa = 11.84AGEE307 pKa = 4.19FDD309 pKa = 3.49WVDD312 pKa = 3.85SVPTDD317 pKa = 3.12QFDD320 pKa = 3.96IIEE323 pKa = 4.58EE324 pKa = 4.2GLPGYY329 pKa = 8.69TILGYY334 pKa = 9.23PDD336 pKa = 3.59RR337 pKa = 11.84GYY339 pKa = 11.38DD340 pKa = 3.51FLGFKK345 pKa = 10.36LGEE348 pKa = 3.81WDD350 pKa = 3.51ADD352 pKa = 3.66EE353 pKa = 6.04GKK355 pKa = 11.13VKK357 pKa = 10.33MNPDD361 pKa = 2.88AKK363 pKa = 10.43MADD366 pKa = 3.42KK367 pKa = 10.86NLRR370 pKa = 11.84QAMAYY375 pKa = 10.22ALDD378 pKa = 4.39LDD380 pKa = 4.02QVAITFSNGLSQRR393 pKa = 11.84ATSHH397 pKa = 5.98IPSNFASFYY406 pKa = 10.48RR407 pKa = 11.84DD408 pKa = 3.49DD409 pKa = 4.44LEE411 pKa = 4.8GYY413 pKa = 9.76PYY415 pKa = 11.09DD416 pKa = 4.35PDD418 pKa = 4.24KK419 pKa = 11.63ANQLLDD425 pKa = 3.38EE426 pKa = 5.28AGYY429 pKa = 11.18EE430 pKa = 4.15DD431 pKa = 4.23VDD433 pKa = 3.67GDD435 pKa = 4.62GFRR438 pKa = 11.84EE439 pKa = 4.48DD440 pKa = 4.64PDD442 pKa = 3.98GQPLEE447 pKa = 3.95ITYY450 pKa = 10.99AAMTGGDD457 pKa = 3.4TAEE460 pKa = 5.52PISQYY465 pKa = 11.15YY466 pKa = 8.2LQAWAEE472 pKa = 4.08VGLNVSLLEE481 pKa = 4.3GRR483 pKa = 11.84LHH485 pKa = 6.84EE486 pKa = 5.11SNSHH490 pKa = 5.34YY491 pKa = 11.12DD492 pKa = 3.91RR493 pKa = 11.84IQGDD497 pKa = 3.82DD498 pKa = 3.59PEE500 pKa = 5.95IDD502 pKa = 3.47VFEE505 pKa = 5.74AGFGALSDD513 pKa = 3.85PTPSVIYY520 pKa = 10.48GPNEE524 pKa = 4.3PINFTRR530 pKa = 11.84YY531 pKa = 7.69EE532 pKa = 3.83TDD534 pKa = 2.93EE535 pKa = 4.2NNEE538 pKa = 3.68YY539 pKa = 10.66LQAFEE544 pKa = 5.0ADD546 pKa = 3.31EE547 pKa = 5.52AFDD550 pKa = 4.72DD551 pKa = 5.64DD552 pKa = 4.3YY553 pKa = 11.5RR554 pKa = 11.84QQLFHH559 pKa = 7.37DD560 pKa = 3.72WQEE563 pKa = 4.04FFMDD567 pKa = 5.62EE568 pKa = 4.4IPTLPTFWFTQLEE581 pKa = 4.41IVNDD585 pKa = 3.82RR586 pKa = 11.84VSAYY590 pKa = 9.81AHH592 pKa = 6.11SSVPQVDD599 pKa = 3.07QDD601 pKa = 3.81TYY603 pKa = 10.76GLHH606 pKa = 6.62EE607 pKa = 4.82IEE609 pKa = 5.32LLAEE613 pKa = 4.39EE614 pKa = 5.22PLTEE618 pKa = 4.0
MM1 pKa = 7.34SLKK4 pKa = 10.27KK5 pKa = 10.25KK6 pKa = 10.21YY7 pKa = 10.59AILASSAALSLTLAACGGEE26 pKa = 4.39GEE28 pKa = 4.66PVDD31 pKa = 4.31PGQEE35 pKa = 4.21GEE37 pKa = 4.39SSQEE41 pKa = 3.93TTEE44 pKa = 4.24DD45 pKa = 2.99QATQEE50 pKa = 4.2RR51 pKa = 11.84SPEE54 pKa = 4.11DD55 pKa = 3.26FDD57 pKa = 3.91EE58 pKa = 4.65TVQNEE63 pKa = 4.22GEE65 pKa = 4.56AIDD68 pKa = 4.76GGIAQVGVLSDD79 pKa = 3.79TAWTGIFDD87 pKa = 3.64WQHH90 pKa = 5.73YY91 pKa = 8.15EE92 pKa = 4.0NEE94 pKa = 4.13ADD96 pKa = 3.58ASLMNYY102 pKa = 8.38TLGDD106 pKa = 4.06LLAMNEE112 pKa = 3.95NYY114 pKa = 10.37EE115 pKa = 4.18LVGGKK120 pKa = 9.85EE121 pKa = 3.71NGAAADD127 pKa = 3.67IEE129 pKa = 4.22FDD131 pKa = 3.77EE132 pKa = 6.23DD133 pKa = 3.79NDD135 pKa = 4.26TVTITVHH142 pKa = 7.51DD143 pKa = 4.04DD144 pKa = 3.96VYY146 pKa = 10.79WHH148 pKa = 7.5DD149 pKa = 4.24GEE151 pKa = 5.39QVTADD156 pKa = 4.35DD157 pKa = 3.86IVFAHH162 pKa = 7.02EE163 pKa = 4.04IVGNPDD169 pKa = 3.1YY170 pKa = 11.26TGIRR174 pKa = 11.84YY175 pKa = 9.58GDD177 pKa = 3.67NFTNVQGMEE186 pKa = 4.07EE187 pKa = 4.41YY188 pKa = 10.52KK189 pKa = 10.84AGEE192 pKa = 4.13ADD194 pKa = 3.92SISGLEE200 pKa = 4.07LSDD203 pKa = 5.85DD204 pKa = 3.69EE205 pKa = 4.59MTVTIHH211 pKa = 5.42YY212 pKa = 8.21QQVVPSMKK220 pKa = 9.55QAGRR224 pKa = 11.84GVWAYY229 pKa = 9.69AAPRR233 pKa = 11.84HH234 pKa = 5.1YY235 pKa = 11.03LEE237 pKa = 6.27DD238 pKa = 3.56IPADD242 pKa = 3.58EE243 pKa = 5.05LVSHH247 pKa = 6.65PNVRR251 pKa = 11.84EE252 pKa = 3.82NPIGFGPFKK261 pKa = 10.59VDD263 pKa = 2.93NVVPGEE269 pKa = 4.17SVEE272 pKa = 3.93YY273 pKa = 10.1SAYY276 pKa = 10.05EE277 pKa = 4.28DD278 pKa = 3.69YY279 pKa = 11.23HH280 pKa = 8.16LGEE283 pKa = 5.1PKK285 pKa = 10.43LDD287 pKa = 4.61GITLEE292 pKa = 4.13RR293 pKa = 11.84LQPAGAIEE301 pKa = 4.03ALRR304 pKa = 11.84AGEE307 pKa = 4.19FDD309 pKa = 3.49WVDD312 pKa = 3.85SVPTDD317 pKa = 3.12QFDD320 pKa = 3.96IIEE323 pKa = 4.58EE324 pKa = 4.2GLPGYY329 pKa = 8.69TILGYY334 pKa = 9.23PDD336 pKa = 3.59RR337 pKa = 11.84GYY339 pKa = 11.38DD340 pKa = 3.51FLGFKK345 pKa = 10.36LGEE348 pKa = 3.81WDD350 pKa = 3.51ADD352 pKa = 3.66EE353 pKa = 6.04GKK355 pKa = 11.13VKK357 pKa = 10.33MNPDD361 pKa = 2.88AKK363 pKa = 10.43MADD366 pKa = 3.42KK367 pKa = 10.86NLRR370 pKa = 11.84QAMAYY375 pKa = 10.22ALDD378 pKa = 4.39LDD380 pKa = 4.02QVAITFSNGLSQRR393 pKa = 11.84ATSHH397 pKa = 5.98IPSNFASFYY406 pKa = 10.48RR407 pKa = 11.84DD408 pKa = 3.49DD409 pKa = 4.44LEE411 pKa = 4.8GYY413 pKa = 9.76PYY415 pKa = 11.09DD416 pKa = 4.35PDD418 pKa = 4.24KK419 pKa = 11.63ANQLLDD425 pKa = 3.38EE426 pKa = 5.28AGYY429 pKa = 11.18EE430 pKa = 4.15DD431 pKa = 4.23VDD433 pKa = 3.67GDD435 pKa = 4.62GFRR438 pKa = 11.84EE439 pKa = 4.48DD440 pKa = 4.64PDD442 pKa = 3.98GQPLEE447 pKa = 3.95ITYY450 pKa = 10.99AAMTGGDD457 pKa = 3.4TAEE460 pKa = 5.52PISQYY465 pKa = 11.15YY466 pKa = 8.2LQAWAEE472 pKa = 4.08VGLNVSLLEE481 pKa = 4.3GRR483 pKa = 11.84LHH485 pKa = 6.84EE486 pKa = 5.11SNSHH490 pKa = 5.34YY491 pKa = 11.12DD492 pKa = 3.91RR493 pKa = 11.84IQGDD497 pKa = 3.82DD498 pKa = 3.59PEE500 pKa = 5.95IDD502 pKa = 3.47VFEE505 pKa = 5.74AGFGALSDD513 pKa = 3.85PTPSVIYY520 pKa = 10.48GPNEE524 pKa = 4.3PINFTRR530 pKa = 11.84YY531 pKa = 7.69EE532 pKa = 3.83TDD534 pKa = 2.93EE535 pKa = 4.2NNEE538 pKa = 3.68YY539 pKa = 10.66LQAFEE544 pKa = 5.0ADD546 pKa = 3.31EE547 pKa = 5.52AFDD550 pKa = 4.72DD551 pKa = 5.64DD552 pKa = 4.3YY553 pKa = 11.5RR554 pKa = 11.84QQLFHH559 pKa = 7.37DD560 pKa = 3.72WQEE563 pKa = 4.04FFMDD567 pKa = 5.62EE568 pKa = 4.4IPTLPTFWFTQLEE581 pKa = 4.41IVNDD585 pKa = 3.82RR586 pKa = 11.84VSAYY590 pKa = 9.81AHH592 pKa = 6.11SSVPQVDD599 pKa = 3.07QDD601 pKa = 3.81TYY603 pKa = 10.76GLHH606 pKa = 6.62EE607 pKa = 4.82IEE609 pKa = 5.32LLAEE613 pKa = 4.39EE614 pKa = 5.22PLTEE618 pKa = 4.0
Molecular weight: 68.74 kDa
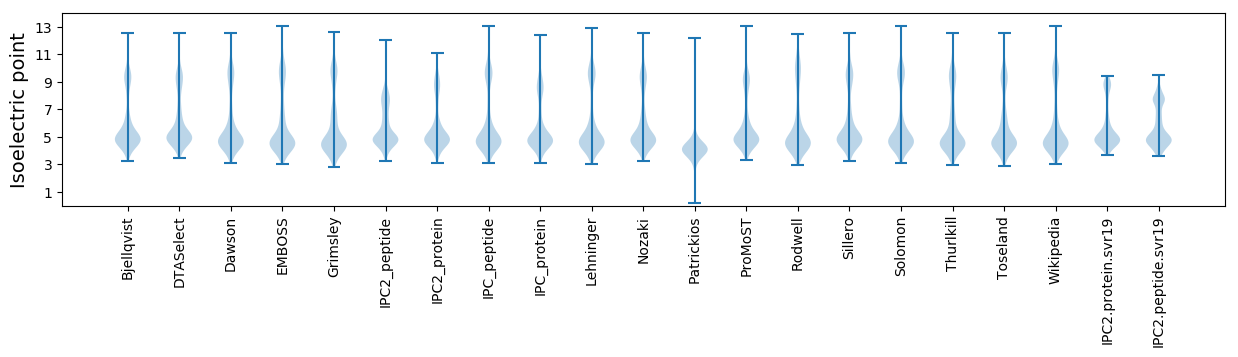
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9EAN6|K9EAN6_9LACT Uncharacterized protein OS=Alloiococcus otitis ATCC 51267 OX=883081 GN=HMPREF9698_00331 PE=4 SV=1
MM1 pKa = 7.42AQGLTFNPKK10 pKa = 9.07KK11 pKa = 10.58RR12 pKa = 11.84KK13 pKa = 8.92GKK15 pKa = 9.67KK16 pKa = 7.8NHH18 pKa = 5.53GFRR21 pKa = 11.84KK22 pKa = 9.98RR23 pKa = 11.84MSTKK27 pKa = 10.18NGRR30 pKa = 11.84KK31 pKa = 8.49ILKK34 pKa = 8.57NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.72GRR41 pKa = 11.84KK42 pKa = 7.92RR43 pKa = 11.84LSYY46 pKa = 11.02
MM1 pKa = 7.42AQGLTFNPKK10 pKa = 9.07KK11 pKa = 10.58RR12 pKa = 11.84KK13 pKa = 8.92GKK15 pKa = 9.67KK16 pKa = 7.8NHH18 pKa = 5.53GFRR21 pKa = 11.84KK22 pKa = 9.98RR23 pKa = 11.84MSTKK27 pKa = 10.18NGRR30 pKa = 11.84KK31 pKa = 8.49ILKK34 pKa = 8.57NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.72GRR41 pKa = 11.84KK42 pKa = 7.92RR43 pKa = 11.84LSYY46 pKa = 11.02
Molecular weight: 5.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
512849 |
35 |
2489 |
315.2 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.365 ± 0.051 | 0.464 ± 0.014 |
7.267 ± 0.065 | 6.927 ± 0.083 |
4.186 ± 0.05 | 6.919 ± 0.054 |
1.852 ± 0.029 | 7.028 ± 0.068 |
5.935 ± 0.071 | 10.154 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.25 ± 0.029 | 4.236 ± 0.039 |
3.584 ± 0.041 | 5.304 ± 0.054 |
4.073 ± 0.051 | 6.227 ± 0.043 |
4.945 ± 0.036 | 6.865 ± 0.048 |
0.804 ± 0.02 | 3.616 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
