
Virgibacillus massiliensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Virgibacillus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
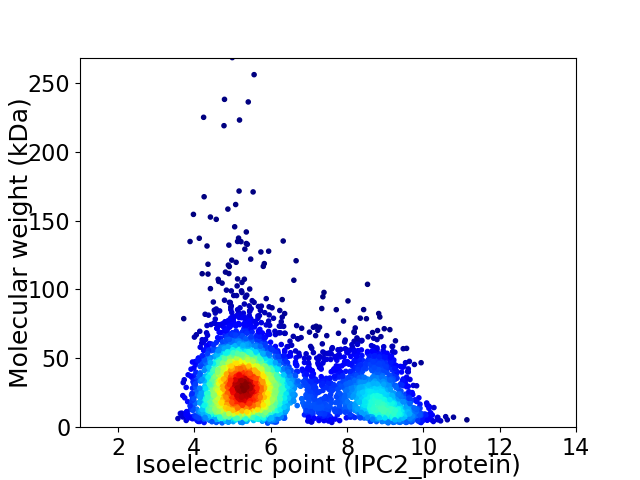
Virtual 2D-PAGE plot for 4389 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024QGD6|A0A024QGD6_9BACI Uncharacterized protein OS=Virgibacillus massiliensis OX=1462526 GN=BN990_03987 PE=4 SV=1
MM1 pKa = 7.8LAQTNHH7 pKa = 6.88KK8 pKa = 9.16ARR10 pKa = 11.84KK11 pKa = 8.37YY12 pKa = 8.82VLSTAFVTSLALTPIFSGSVFANAGAGASEE42 pKa = 4.33GDD44 pKa = 3.69LTISEE49 pKa = 4.55SNIPNVEE56 pKa = 4.0QQEE59 pKa = 4.43DD60 pKa = 4.0TSSNEE65 pKa = 3.09ASLIQRR71 pKa = 11.84GDD73 pKa = 3.24VSEE76 pKa = 4.36SVEE79 pKa = 4.16DD80 pKa = 3.94VQALLQDD87 pKa = 3.36QGYY90 pKa = 7.75YY91 pKa = 10.03TYY93 pKa = 10.35TVDD96 pKa = 5.68GIFGPITEE104 pKa = 4.27QAVRR108 pKa = 11.84DD109 pKa = 3.92YY110 pKa = 11.21QADD113 pKa = 2.97QGLQVDD119 pKa = 5.6GIVGPNTSEE128 pKa = 3.98AMGLSGSEE136 pKa = 4.1SSNDD140 pKa = 3.32LTITEE145 pKa = 4.79SGDD148 pKa = 3.43NSDD151 pKa = 3.32IQADD155 pKa = 3.32IVAAAEE161 pKa = 4.32SVIGTPYY168 pKa = 10.07VWGGTTTDD176 pKa = 3.88GMDD179 pKa = 2.84SSGFINYY186 pKa = 8.91VFDD189 pKa = 3.56QVGIDD194 pKa = 3.34VSRR197 pKa = 11.84THH199 pKa = 8.37SEE201 pKa = 3.18MWQNDD206 pKa = 4.08GVHH209 pKa = 6.22VDD211 pKa = 3.65APSVGDD217 pKa = 3.45VVFFEE222 pKa = 4.58GTYY225 pKa = 7.29EE226 pKa = 4.1TEE228 pKa = 4.4GASHH232 pKa = 6.3SGIYY236 pKa = 10.04IGDD239 pKa = 3.51NQMIHH244 pKa = 6.87AGNDD248 pKa = 3.66GVSVADD254 pKa = 4.07ISIDD258 pKa = 3.29YY259 pKa = 8.08WKK261 pKa = 10.54NHH263 pKa = 6.04YY264 pKa = 9.97IGAKK268 pKa = 9.94SITEE272 pKa = 3.89
MM1 pKa = 7.8LAQTNHH7 pKa = 6.88KK8 pKa = 9.16ARR10 pKa = 11.84KK11 pKa = 8.37YY12 pKa = 8.82VLSTAFVTSLALTPIFSGSVFANAGAGASEE42 pKa = 4.33GDD44 pKa = 3.69LTISEE49 pKa = 4.55SNIPNVEE56 pKa = 4.0QQEE59 pKa = 4.43DD60 pKa = 4.0TSSNEE65 pKa = 3.09ASLIQRR71 pKa = 11.84GDD73 pKa = 3.24VSEE76 pKa = 4.36SVEE79 pKa = 4.16DD80 pKa = 3.94VQALLQDD87 pKa = 3.36QGYY90 pKa = 7.75YY91 pKa = 10.03TYY93 pKa = 10.35TVDD96 pKa = 5.68GIFGPITEE104 pKa = 4.27QAVRR108 pKa = 11.84DD109 pKa = 3.92YY110 pKa = 11.21QADD113 pKa = 2.97QGLQVDD119 pKa = 5.6GIVGPNTSEE128 pKa = 3.98AMGLSGSEE136 pKa = 4.1SSNDD140 pKa = 3.32LTITEE145 pKa = 4.79SGDD148 pKa = 3.43NSDD151 pKa = 3.32IQADD155 pKa = 3.32IVAAAEE161 pKa = 4.32SVIGTPYY168 pKa = 10.07VWGGTTTDD176 pKa = 3.88GMDD179 pKa = 2.84SSGFINYY186 pKa = 8.91VFDD189 pKa = 3.56QVGIDD194 pKa = 3.34VSRR197 pKa = 11.84THH199 pKa = 8.37SEE201 pKa = 3.18MWQNDD206 pKa = 4.08GVHH209 pKa = 6.22VDD211 pKa = 3.65APSVGDD217 pKa = 3.45VVFFEE222 pKa = 4.58GTYY225 pKa = 7.29EE226 pKa = 4.1TEE228 pKa = 4.4GASHH232 pKa = 6.3SGIYY236 pKa = 10.04IGDD239 pKa = 3.51NQMIHH244 pKa = 6.87AGNDD248 pKa = 3.66GVSVADD254 pKa = 4.07ISIDD258 pKa = 3.29YY259 pKa = 8.08WKK261 pKa = 10.54NHH263 pKa = 6.04YY264 pKa = 9.97IGAKK268 pKa = 9.94SITEE272 pKa = 3.89
Molecular weight: 28.85 kDa
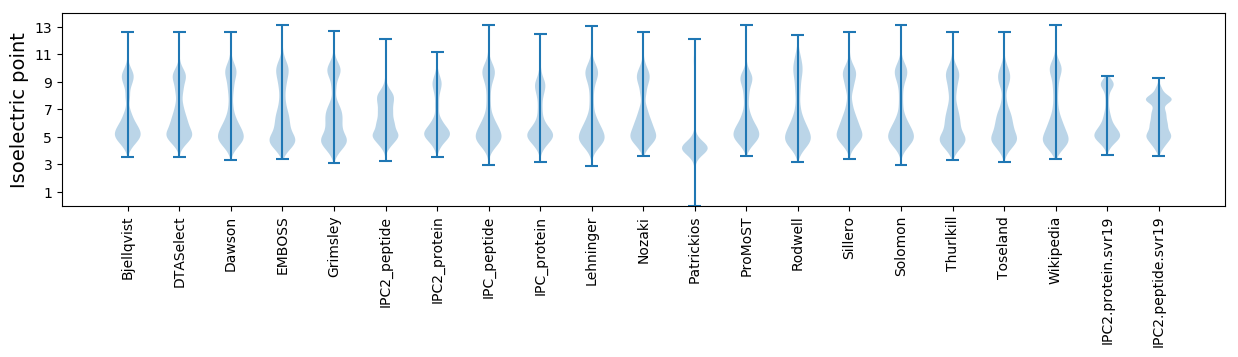
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024Q6D6|A0A024Q6D6_9BACI Ribosomal-protein-alanine N-acetyltransferase OS=Virgibacillus massiliensis OX=1462526 GN=BN990_00355 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.35QPNNRR10 pKa = 11.84KK11 pKa = 9.22RR12 pKa = 11.84KK13 pKa = 8.02KK14 pKa = 8.47VHH16 pKa = 5.57GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 10.29NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.35QPNNRR10 pKa = 11.84KK11 pKa = 9.22RR12 pKa = 11.84KK13 pKa = 8.02KK14 pKa = 8.47VHH16 pKa = 5.57GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 10.29NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1246616 |
29 |
2377 |
284.0 |
31.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.959 ± 0.035 | 0.674 ± 0.011 |
5.289 ± 0.035 | 7.207 ± 0.046 |
4.496 ± 0.033 | 6.753 ± 0.038 |
2.2 ± 0.022 | 8.289 ± 0.044 |
6.505 ± 0.041 | 9.416 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.848 ± 0.018 | 4.776 ± 0.034 |
3.578 ± 0.025 | 4.116 ± 0.032 |
3.832 ± 0.029 | 6.0 ± 0.031 |
5.637 ± 0.024 | 6.669 ± 0.032 |
1.073 ± 0.016 | 3.682 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
