
Micromonas pusilla (strain CCMP1545) (Picoplanktonic green alga)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas; Micromonas pusilla
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
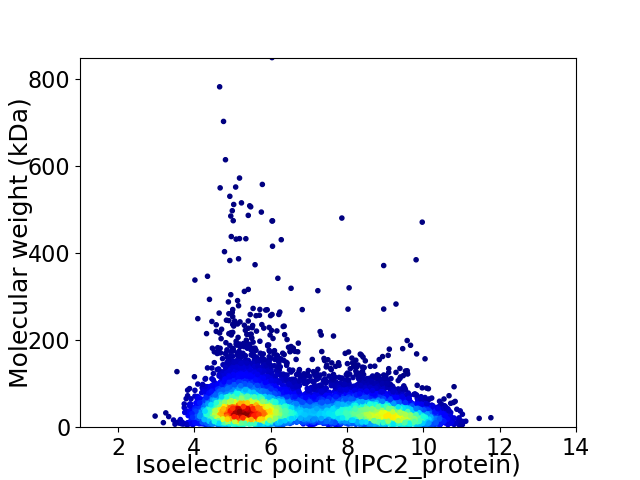
Virtual 2D-PAGE plot for 10250 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C1MWV0|C1MWV0_MICPC Predicted protein OS=Micromonas pusilla (strain CCMP1545) OX=564608 GN=MICPUCDRAFT_59262 PE=4 SV=1
MM1 pKa = 6.75TRR3 pKa = 11.84GRR5 pKa = 11.84ARR7 pKa = 11.84DD8 pKa = 3.38AAAAAAAAARR18 pKa = 11.84VAVLLLLLLLARR30 pKa = 11.84AASAQSPPPRR40 pKa = 11.84SRR42 pKa = 11.84SPAFGSCVDD51 pKa = 3.43ACGSCWYY58 pKa = 10.16EE59 pKa = 3.62RR60 pKa = 11.84DD61 pKa = 3.5DD62 pKa = 3.94PSRR65 pKa = 11.84VSCCCEE71 pKa = 3.87PTCADD76 pKa = 5.5AGDD79 pKa = 4.37CCDD82 pKa = 5.68DD83 pKa = 3.63YY84 pKa = 12.28AEE86 pKa = 4.38ACEE89 pKa = 4.84ANGDD93 pKa = 3.99DD94 pKa = 4.12EE95 pKa = 6.11RR96 pKa = 11.84GGDD99 pKa = 3.87GEE101 pKa = 4.43RR102 pKa = 11.84GVAGVVAEE110 pKa = 4.45ANGDD114 pKa = 3.56AALFFAPAPGPGASALFAAPIAVDD138 pKa = 3.78VEE140 pKa = 4.43DD141 pKa = 5.71DD142 pKa = 4.22ADD144 pKa = 4.18DD145 pKa = 4.63DD146 pKa = 4.66APVRR150 pKa = 11.84APDD153 pKa = 4.21PPTAAPAPSPSSVAYY168 pKa = 8.8FVAPAEE174 pKa = 4.16EE175 pKa = 4.33EE176 pKa = 4.21EE177 pKa = 4.43EE178 pKa = 4.27EE179 pKa = 4.68EE180 pKa = 4.21EE181 pKa = 4.56TEE183 pKa = 4.11TDD185 pKa = 3.07ATAAPPPTPRR195 pKa = 11.84VVPTIPEE202 pKa = 4.2EE203 pKa = 4.22PSPSPPSPSPPSPSPPSPSPPPPSPSPPPIAPPPPVVEE241 pKa = 4.4EE242 pKa = 4.35GVVADD247 pKa = 4.78VVEE250 pKa = 4.53EE251 pKa = 4.1DD252 pKa = 5.71DD253 pKa = 5.82GYY255 pKa = 11.81QNQNQNDD262 pKa = 3.96YY263 pKa = 11.68DD264 pKa = 5.58DD265 pKa = 6.41DD266 pKa = 7.22DD267 pKa = 7.47DD268 pKa = 7.66DD269 pKa = 7.62DD270 pKa = 7.01DD271 pKa = 7.22DD272 pKa = 5.72EE273 pKa = 7.46DD274 pKa = 4.65DD275 pKa = 4.3AVLVDD280 pKa = 3.64VSGGAGGVLPTAKK293 pKa = 9.89FWEE296 pKa = 4.64AVADD300 pKa = 4.42RR301 pKa = 11.84LGLDD305 pKa = 4.2LRR307 pKa = 11.84GNPAKK312 pKa = 10.53EE313 pKa = 4.07EE314 pKa = 3.96EE315 pKa = 4.5SEE317 pKa = 4.28LEE319 pKa = 3.95QEE321 pKa = 4.21EE322 pKa = 4.72EE323 pKa = 3.84EE324 pKa = 4.99DD325 pKa = 4.01EE326 pKa = 4.27EE327 pKa = 5.24EE328 pKa = 4.2KK329 pKa = 10.63TKK331 pKa = 11.28DD332 pKa = 3.14DD333 pKa = 3.72APRR336 pKa = 11.84VGGGGPPKK344 pKa = 10.51ASTGRR349 pKa = 11.84RR350 pKa = 11.84DD351 pKa = 5.35DD352 pKa = 5.52DD353 pKa = 6.11DD354 pKa = 7.14DD355 pKa = 4.31DD356 pKa = 5.76ANPISTAGEE365 pKa = 3.8NAFCLDD371 pKa = 3.48KK372 pKa = 11.51SEE374 pKa = 3.98EE375 pKa = 4.04RR376 pKa = 11.84YY377 pKa = 10.11YY378 pKa = 11.55ADD380 pKa = 2.97TTRR383 pKa = 11.84ACDD386 pKa = 4.19CDD388 pKa = 4.9CAGCDD393 pKa = 3.29CRR395 pKa = 11.84RR396 pKa = 11.84YY397 pKa = 8.59YY398 pKa = 11.09ACWDD402 pKa = 3.72CDD404 pKa = 3.35GGACASQASYY414 pKa = 10.78EE415 pKa = 4.39CARR418 pKa = 11.84GDD420 pKa = 3.23YY421 pKa = 10.01STFNEE426 pKa = 4.38KK427 pKa = 10.55LQLCAWEE434 pKa = 4.38QPRR437 pKa = 11.84PLPFSCPPRR446 pKa = 11.84PPGPPNPPNPPRR458 pKa = 11.84PPPRR462 pKa = 11.84PAAPPPLPGFPAAPPNPPALAPLPTPLPQPTLQPGVVPGGGVGGFPGGGVAQPGLGGGVVGGVPGAGAGVGQPGAGVGQPSQFQPGFGAQPGFGAQPGFGAQPGGAVSPVQTQPGSCASIPQCNSCLQSSDD593 pKa = 3.03QSSFVCCCDD602 pKa = 3.95PDD604 pKa = 4.28CSTWNANAPGQPAGFLGCCSDD625 pKa = 3.8YY626 pKa = 11.48AATCGAGQAIPGRR639 pKa = 11.84QQRR642 pKa = 11.84GLLL645 pKa = 3.72
MM1 pKa = 6.75TRR3 pKa = 11.84GRR5 pKa = 11.84ARR7 pKa = 11.84DD8 pKa = 3.38AAAAAAAAARR18 pKa = 11.84VAVLLLLLLLARR30 pKa = 11.84AASAQSPPPRR40 pKa = 11.84SRR42 pKa = 11.84SPAFGSCVDD51 pKa = 3.43ACGSCWYY58 pKa = 10.16EE59 pKa = 3.62RR60 pKa = 11.84DD61 pKa = 3.5DD62 pKa = 3.94PSRR65 pKa = 11.84VSCCCEE71 pKa = 3.87PTCADD76 pKa = 5.5AGDD79 pKa = 4.37CCDD82 pKa = 5.68DD83 pKa = 3.63YY84 pKa = 12.28AEE86 pKa = 4.38ACEE89 pKa = 4.84ANGDD93 pKa = 3.99DD94 pKa = 4.12EE95 pKa = 6.11RR96 pKa = 11.84GGDD99 pKa = 3.87GEE101 pKa = 4.43RR102 pKa = 11.84GVAGVVAEE110 pKa = 4.45ANGDD114 pKa = 3.56AALFFAPAPGPGASALFAAPIAVDD138 pKa = 3.78VEE140 pKa = 4.43DD141 pKa = 5.71DD142 pKa = 4.22ADD144 pKa = 4.18DD145 pKa = 4.63DD146 pKa = 4.66APVRR150 pKa = 11.84APDD153 pKa = 4.21PPTAAPAPSPSSVAYY168 pKa = 8.8FVAPAEE174 pKa = 4.16EE175 pKa = 4.33EE176 pKa = 4.21EE177 pKa = 4.43EE178 pKa = 4.27EE179 pKa = 4.68EE180 pKa = 4.21EE181 pKa = 4.56TEE183 pKa = 4.11TDD185 pKa = 3.07ATAAPPPTPRR195 pKa = 11.84VVPTIPEE202 pKa = 4.2EE203 pKa = 4.22PSPSPPSPSPPSPSPPSPSPPPPSPSPPPIAPPPPVVEE241 pKa = 4.4EE242 pKa = 4.35GVVADD247 pKa = 4.78VVEE250 pKa = 4.53EE251 pKa = 4.1DD252 pKa = 5.71DD253 pKa = 5.82GYY255 pKa = 11.81QNQNQNDD262 pKa = 3.96YY263 pKa = 11.68DD264 pKa = 5.58DD265 pKa = 6.41DD266 pKa = 7.22DD267 pKa = 7.47DD268 pKa = 7.66DD269 pKa = 7.62DD270 pKa = 7.01DD271 pKa = 7.22DD272 pKa = 5.72EE273 pKa = 7.46DD274 pKa = 4.65DD275 pKa = 4.3AVLVDD280 pKa = 3.64VSGGAGGVLPTAKK293 pKa = 9.89FWEE296 pKa = 4.64AVADD300 pKa = 4.42RR301 pKa = 11.84LGLDD305 pKa = 4.2LRR307 pKa = 11.84GNPAKK312 pKa = 10.53EE313 pKa = 4.07EE314 pKa = 3.96EE315 pKa = 4.5SEE317 pKa = 4.28LEE319 pKa = 3.95QEE321 pKa = 4.21EE322 pKa = 4.72EE323 pKa = 3.84EE324 pKa = 4.99DD325 pKa = 4.01EE326 pKa = 4.27EE327 pKa = 5.24EE328 pKa = 4.2KK329 pKa = 10.63TKK331 pKa = 11.28DD332 pKa = 3.14DD333 pKa = 3.72APRR336 pKa = 11.84VGGGGPPKK344 pKa = 10.51ASTGRR349 pKa = 11.84RR350 pKa = 11.84DD351 pKa = 5.35DD352 pKa = 5.52DD353 pKa = 6.11DD354 pKa = 7.14DD355 pKa = 4.31DD356 pKa = 5.76ANPISTAGEE365 pKa = 3.8NAFCLDD371 pKa = 3.48KK372 pKa = 11.51SEE374 pKa = 3.98EE375 pKa = 4.04RR376 pKa = 11.84YY377 pKa = 10.11YY378 pKa = 11.55ADD380 pKa = 2.97TTRR383 pKa = 11.84ACDD386 pKa = 4.19CDD388 pKa = 4.9CAGCDD393 pKa = 3.29CRR395 pKa = 11.84RR396 pKa = 11.84YY397 pKa = 8.59YY398 pKa = 11.09ACWDD402 pKa = 3.72CDD404 pKa = 3.35GGACASQASYY414 pKa = 10.78EE415 pKa = 4.39CARR418 pKa = 11.84GDD420 pKa = 3.23YY421 pKa = 10.01STFNEE426 pKa = 4.38KK427 pKa = 10.55LQLCAWEE434 pKa = 4.38QPRR437 pKa = 11.84PLPFSCPPRR446 pKa = 11.84PPGPPNPPNPPRR458 pKa = 11.84PPPRR462 pKa = 11.84PAAPPPLPGFPAAPPNPPALAPLPTPLPQPTLQPGVVPGGGVGGFPGGGVAQPGLGGGVVGGVPGAGAGVGQPGAGVGQPSQFQPGFGAQPGFGAQPGFGAQPGGAVSPVQTQPGSCASIPQCNSCLQSSDD593 pKa = 3.03QSSFVCCCDD602 pKa = 3.95PDD604 pKa = 4.28CSTWNANAPGQPAGFLGCCSDD625 pKa = 3.8YY626 pKa = 11.48AATCGAGQAIPGRR639 pKa = 11.84QQRR642 pKa = 11.84GLLL645 pKa = 3.72
Molecular weight: 65.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C1N6Q0|C1N6Q0_MICPC Predicted protein OS=Micromonas pusilla (strain CCMP1545) OX=564608 GN=MICPUCDRAFT_53390 PE=4 SV=1
MM1 pKa = 6.94TTTTTTTTTRR11 pKa = 11.84RR12 pKa = 11.84TKK14 pKa = 8.58RR15 pKa = 11.84TKK17 pKa = 10.47RR18 pKa = 11.84RR19 pKa = 11.84TKK21 pKa = 9.07RR22 pKa = 11.84TKK24 pKa = 8.66TRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84FKK31 pKa = 10.23PRR33 pKa = 11.84TRR35 pKa = 11.84TRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84SRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84PALGLLKK59 pKa = 10.32RR60 pKa = 11.84RR61 pKa = 11.84SRR63 pKa = 11.84LAIAPRR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84TSTFRR77 pKa = 11.84GARR80 pKa = 11.84WRR82 pKa = 11.84RR83 pKa = 11.84CSPRR87 pKa = 11.84WATRR91 pKa = 11.84SRR93 pKa = 11.84SSRR96 pKa = 11.84PRR98 pKa = 11.84RR99 pKa = 11.84SRR101 pKa = 11.84ARR103 pKa = 11.84ARR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84PTLPCRR127 pKa = 11.84TRR129 pKa = 11.84CRR131 pKa = 11.84TRR133 pKa = 11.84PMTRR137 pKa = 11.84LATRR141 pKa = 11.84WRR143 pKa = 11.84PRR145 pKa = 11.84PPRR148 pKa = 11.84WTRR151 pKa = 11.84STPRR155 pKa = 11.84SPRR158 pKa = 11.84SS159 pKa = 3.19
MM1 pKa = 6.94TTTTTTTTTRR11 pKa = 11.84RR12 pKa = 11.84TKK14 pKa = 8.58RR15 pKa = 11.84TKK17 pKa = 10.47RR18 pKa = 11.84RR19 pKa = 11.84TKK21 pKa = 9.07RR22 pKa = 11.84TKK24 pKa = 8.66TRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84FKK31 pKa = 10.23PRR33 pKa = 11.84TRR35 pKa = 11.84TRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84SRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84PALGLLKK59 pKa = 10.32RR60 pKa = 11.84RR61 pKa = 11.84SRR63 pKa = 11.84LAIAPRR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84TSTFRR77 pKa = 11.84GARR80 pKa = 11.84WRR82 pKa = 11.84RR83 pKa = 11.84CSPRR87 pKa = 11.84WATRR91 pKa = 11.84SRR93 pKa = 11.84SSRR96 pKa = 11.84PRR98 pKa = 11.84RR99 pKa = 11.84SRR101 pKa = 11.84ARR103 pKa = 11.84ARR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84PTLPCRR127 pKa = 11.84TRR129 pKa = 11.84CRR131 pKa = 11.84TRR133 pKa = 11.84PMTRR137 pKa = 11.84LATRR141 pKa = 11.84WRR143 pKa = 11.84PRR145 pKa = 11.84PPRR148 pKa = 11.84WTRR151 pKa = 11.84STPRR155 pKa = 11.84SPRR158 pKa = 11.84SS159 pKa = 3.19
Molecular weight: 20.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4600988 |
35 |
7926 |
448.9 |
48.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.193 ± 0.064 | 1.431 ± 0.012 |
6.678 ± 0.023 | 6.499 ± 0.029 |
3.094 ± 0.015 | 8.59 ± 0.036 |
1.951 ± 0.012 | 2.987 ± 0.017 |
4.354 ± 0.03 | 7.707 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.99 ± 0.012 | 2.494 ± 0.014 |
5.48 ± 0.027 | 2.069 ± 0.016 |
8.129 ± 0.033 | 6.74 ± 0.025 |
5.341 ± 0.019 | 7.12 ± 0.023 |
1.208 ± 0.01 | 1.942 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
