
Gyrovirus GyV8
Taxonomy: Viruses; Anelloviridae; Gyrovirus; Gyrovirus fulgla1
Average proteome isoelectric point is 8.0
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G4ANJ1|A0A0G4ANJ1_9VIRU CA1 OS=Gyrovirus GyV8 OX=1670973 PE=3 SV=1
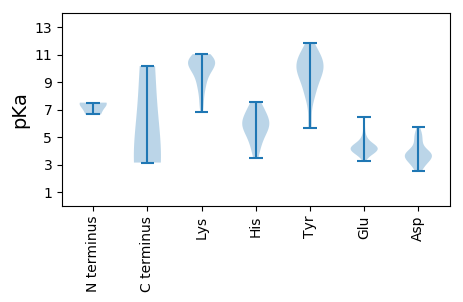
MM1 pKa = 6.64QHH3 pKa = 5.73VARR6 pKa = 11.84SDD8 pKa = 3.18VSMEE12 pKa = 4.27YY13 pKa = 9.9ILEE16 pKa = 3.92PTRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84AEE24 pKa = 3.74QNKK27 pKa = 9.68QIAKK31 pKa = 9.29WYY33 pKa = 8.59RR34 pKa = 11.84EE35 pKa = 3.86CSRR38 pKa = 11.84LHH40 pKa = 6.96RR41 pKa = 11.84NYY43 pKa = 8.56CTCSSWQSHH52 pKa = 5.46FRR54 pKa = 11.84VQKK57 pKa = 9.58PEE59 pKa = 3.81KK60 pKa = 10.2KK61 pKa = 9.98NVSTQTQRR69 pKa = 11.84TDD71 pKa = 3.07PSVEE75 pKa = 3.29IGYY78 pKa = 9.67IPTGHH83 pKa = 7.58PLFKK87 pKa = 10.7SADD90 pKa = 3.81RR91 pKa = 11.84ARR93 pKa = 11.84SKK95 pKa = 11.03GLRR98 pKa = 11.84SYY100 pKa = 11.15SIDD103 pKa = 3.54PFLSPPFTQAAEE115 pKa = 3.95GRR117 pKa = 11.84TGKK120 pKa = 10.25RR121 pKa = 11.84KK122 pKa = 9.61RR123 pKa = 11.84ARR125 pKa = 11.84KK126 pKa = 8.53QDD128 pKa = 3.7LYY130 pKa = 11.24CRR132 pKa = 11.84EE133 pKa = 4.16EE134 pKa = 5.09LSDD137 pKa = 4.61DD138 pKa = 5.16DD139 pKa = 5.71YY140 pKa = 11.81NTTPKK145 pKa = 10.21QLRR148 pKa = 11.84AWLGDD153 pKa = 3.58LGEE156 pKa = 4.74DD157 pKa = 3.41TTTSEE162 pKa = 4.43EE163 pKa = 4.35ADD165 pKa = 3.61GTDD168 pKa = 3.27TDD170 pKa = 4.21GTQDD174 pKa = 3.34TEE176 pKa = 4.93GDD178 pKa = 3.68MEE180 pKa = 6.45DD181 pKa = 4.51DD182 pKa = 3.9FGAAITEE189 pKa = 4.45GNINADD195 pKa = 3.85PLIDD199 pKa = 3.69MLSGSFSTQNLASTPFDD216 pKa = 3.64YY217 pKa = 9.27QTPTTDD223 pKa = 2.57KK224 pKa = 10.56TSFFKK229 pKa = 10.87ASYY232 pKa = 10.16
MM1 pKa = 6.64QHH3 pKa = 5.73VARR6 pKa = 11.84SDD8 pKa = 3.18VSMEE12 pKa = 4.27YY13 pKa = 9.9ILEE16 pKa = 3.92PTRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84AEE24 pKa = 3.74QNKK27 pKa = 9.68QIAKK31 pKa = 9.29WYY33 pKa = 8.59RR34 pKa = 11.84EE35 pKa = 3.86CSRR38 pKa = 11.84LHH40 pKa = 6.96RR41 pKa = 11.84NYY43 pKa = 8.56CTCSSWQSHH52 pKa = 5.46FRR54 pKa = 11.84VQKK57 pKa = 9.58PEE59 pKa = 3.81KK60 pKa = 10.2KK61 pKa = 9.98NVSTQTQRR69 pKa = 11.84TDD71 pKa = 3.07PSVEE75 pKa = 3.29IGYY78 pKa = 9.67IPTGHH83 pKa = 7.58PLFKK87 pKa = 10.7SADD90 pKa = 3.81RR91 pKa = 11.84ARR93 pKa = 11.84SKK95 pKa = 11.03GLRR98 pKa = 11.84SYY100 pKa = 11.15SIDD103 pKa = 3.54PFLSPPFTQAAEE115 pKa = 3.95GRR117 pKa = 11.84TGKK120 pKa = 10.25RR121 pKa = 11.84KK122 pKa = 9.61RR123 pKa = 11.84ARR125 pKa = 11.84KK126 pKa = 8.53QDD128 pKa = 3.7LYY130 pKa = 11.24CRR132 pKa = 11.84EE133 pKa = 4.16EE134 pKa = 5.09LSDD137 pKa = 4.61DD138 pKa = 5.16DD139 pKa = 5.71YY140 pKa = 11.81NTTPKK145 pKa = 10.21QLRR148 pKa = 11.84AWLGDD153 pKa = 3.58LGEE156 pKa = 4.74DD157 pKa = 3.41TTTSEE162 pKa = 4.43EE163 pKa = 4.35ADD165 pKa = 3.61GTDD168 pKa = 3.27TDD170 pKa = 4.21GTQDD174 pKa = 3.34TEE176 pKa = 4.93GDD178 pKa = 3.68MEE180 pKa = 6.45DD181 pKa = 4.51DD182 pKa = 3.9FGAAITEE189 pKa = 4.45GNINADD195 pKa = 3.85PLIDD199 pKa = 3.69MLSGSFSTQNLASTPFDD216 pKa = 3.64YY217 pKa = 9.27QTPTTDD223 pKa = 2.57KK224 pKa = 10.56TSFFKK229 pKa = 10.87ASYY232 pKa = 10.16
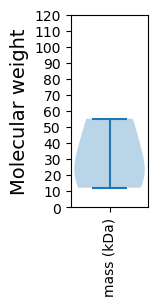
Molecular weight: 26.41 kDa
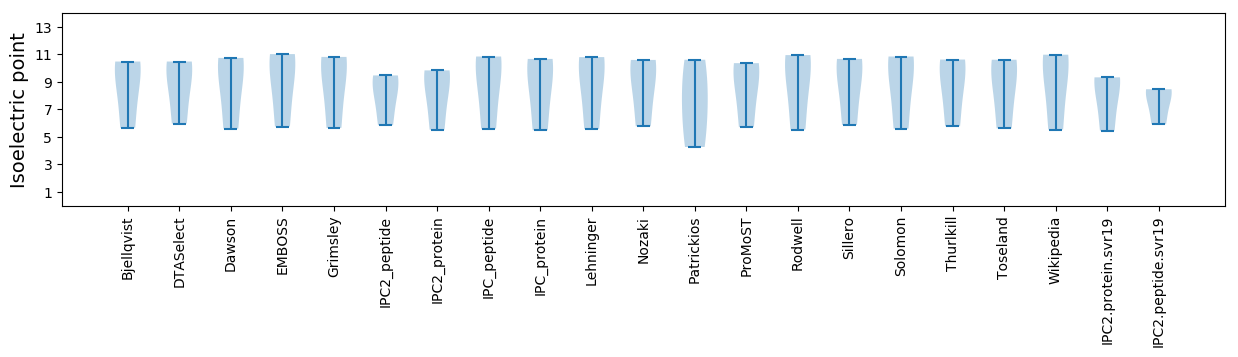
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G4ANJ1|A0A0G4ANJ1_9VIRU CA1 OS=Gyrovirus GyV8 OX=1670973 PE=3 SV=1
MM1 pKa = 7.5ARR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84GRR8 pKa = 11.84YY9 pKa = 5.64YY10 pKa = 10.43HH11 pKa = 6.14FRR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84WHH18 pKa = 5.91RR19 pKa = 11.84HH20 pKa = 3.47RR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.13PRR25 pKa = 11.84HH26 pKa = 5.42RR27 pKa = 11.84RR28 pKa = 11.84GYY30 pKa = 7.97GRR32 pKa = 11.84RR33 pKa = 11.84FRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84HH38 pKa = 4.42YY39 pKa = 10.01RR40 pKa = 11.84GKK42 pKa = 9.93HH43 pKa = 3.97KK44 pKa = 10.32RR45 pKa = 11.84RR46 pKa = 11.84SFNRR50 pKa = 11.84YY51 pKa = 6.29VKK53 pKa = 10.53RR54 pKa = 11.84LFFNPKK60 pKa = 8.48PGFYY64 pKa = 9.36TVRR67 pKa = 11.84LPNPYY72 pKa = 9.59NRR74 pKa = 11.84QNIFFQGLILVPLGVDD90 pKa = 2.97NLSAVSYY97 pKa = 10.25KK98 pKa = 10.71SEE100 pKa = 4.03FCRR103 pKa = 11.84VASITLSLRR112 pKa = 11.84NLLLAVFPLDD122 pKa = 4.74AISKK126 pKa = 9.84AGGPIPDD133 pKa = 5.29IITAYY138 pKa = 8.86HH139 pKa = 6.49TATEE143 pKa = 4.31HH144 pKa = 6.09QSQNLPPWTNMPQKK158 pKa = 10.3GNASATPFDD167 pKa = 3.2WWRR170 pKa = 11.84WSLIMMRR177 pKa = 11.84PEE179 pKa = 4.31DD180 pKa = 3.84NVRR183 pKa = 11.84RR184 pKa = 11.84AQFPYY189 pKa = 9.81FPEE192 pKa = 4.09RR193 pKa = 11.84VEE195 pKa = 5.68DD196 pKa = 3.8LLQILGGWHH205 pKa = 7.47LFRR208 pKa = 11.84HH209 pKa = 6.37IKK211 pKa = 6.81TTMVVMATRR220 pKa = 11.84GMSGFDD226 pKa = 3.24PVASLFVQDD235 pKa = 4.92SYY237 pKa = 11.38WLSRR241 pKa = 11.84TGNPSIINEE250 pKa = 4.07NGNVKK255 pKa = 10.01LQAPMPGGGVKK266 pKa = 10.36GRR268 pKa = 11.84IEE270 pKa = 4.21TSTITGPPCDD280 pKa = 3.84EE281 pKa = 4.15YY282 pKa = 10.69MFPSFPPQWSDD293 pKa = 2.51KK294 pKa = 8.14TQPTQSFKK302 pKa = 10.88QDD304 pKa = 2.77NAFYY308 pKa = 10.93CLACYY313 pKa = 9.84PSFLTLSALGAPWAFPPTEE332 pKa = 4.17KK333 pKa = 10.53SVSRR337 pKa = 11.84SSFNHH342 pKa = 6.18HH343 pKa = 6.38SVTGNNDD350 pKa = 3.2PQGRR354 pKa = 11.84RR355 pKa = 11.84WMTFLPKK362 pKa = 8.92PTALQVPNPTGHH374 pKa = 6.93GEE376 pKa = 4.34TPTSSPGPTGPPPTPTEE393 pKa = 3.65IDD395 pKa = 3.48TPIGVLFLAQTSWGDD410 pKa = 4.01FINHH414 pKa = 4.61TWGTKK419 pKa = 9.18PYY421 pKa = 10.64NFGTTSDD428 pKa = 3.96EE429 pKa = 4.56SPNFPPFSRR438 pKa = 11.84DD439 pKa = 3.08TQNRR443 pKa = 11.84PWAVIKK449 pKa = 10.76LKK451 pKa = 11.03SKK453 pKa = 8.85WQLGNKK459 pKa = 7.5QRR461 pKa = 11.84PYY463 pKa = 10.35WYY465 pKa = 7.78DD466 pKa = 3.61TNWWKK471 pKa = 10.48QSRR474 pKa = 11.84TQFGG478 pKa = 3.14
MM1 pKa = 7.5ARR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84GRR8 pKa = 11.84YY9 pKa = 5.64YY10 pKa = 10.43HH11 pKa = 6.14FRR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84WHH18 pKa = 5.91RR19 pKa = 11.84HH20 pKa = 3.47RR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.13PRR25 pKa = 11.84HH26 pKa = 5.42RR27 pKa = 11.84RR28 pKa = 11.84GYY30 pKa = 7.97GRR32 pKa = 11.84RR33 pKa = 11.84FRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84HH38 pKa = 4.42YY39 pKa = 10.01RR40 pKa = 11.84GKK42 pKa = 9.93HH43 pKa = 3.97KK44 pKa = 10.32RR45 pKa = 11.84RR46 pKa = 11.84SFNRR50 pKa = 11.84YY51 pKa = 6.29VKK53 pKa = 10.53RR54 pKa = 11.84LFFNPKK60 pKa = 8.48PGFYY64 pKa = 9.36TVRR67 pKa = 11.84LPNPYY72 pKa = 9.59NRR74 pKa = 11.84QNIFFQGLILVPLGVDD90 pKa = 2.97NLSAVSYY97 pKa = 10.25KK98 pKa = 10.71SEE100 pKa = 4.03FCRR103 pKa = 11.84VASITLSLRR112 pKa = 11.84NLLLAVFPLDD122 pKa = 4.74AISKK126 pKa = 9.84AGGPIPDD133 pKa = 5.29IITAYY138 pKa = 8.86HH139 pKa = 6.49TATEE143 pKa = 4.31HH144 pKa = 6.09QSQNLPPWTNMPQKK158 pKa = 10.3GNASATPFDD167 pKa = 3.2WWRR170 pKa = 11.84WSLIMMRR177 pKa = 11.84PEE179 pKa = 4.31DD180 pKa = 3.84NVRR183 pKa = 11.84RR184 pKa = 11.84AQFPYY189 pKa = 9.81FPEE192 pKa = 4.09RR193 pKa = 11.84VEE195 pKa = 5.68DD196 pKa = 3.8LLQILGGWHH205 pKa = 7.47LFRR208 pKa = 11.84HH209 pKa = 6.37IKK211 pKa = 6.81TTMVVMATRR220 pKa = 11.84GMSGFDD226 pKa = 3.24PVASLFVQDD235 pKa = 4.92SYY237 pKa = 11.38WLSRR241 pKa = 11.84TGNPSIINEE250 pKa = 4.07NGNVKK255 pKa = 10.01LQAPMPGGGVKK266 pKa = 10.36GRR268 pKa = 11.84IEE270 pKa = 4.21TSTITGPPCDD280 pKa = 3.84EE281 pKa = 4.15YY282 pKa = 10.69MFPSFPPQWSDD293 pKa = 2.51KK294 pKa = 8.14TQPTQSFKK302 pKa = 10.88QDD304 pKa = 2.77NAFYY308 pKa = 10.93CLACYY313 pKa = 9.84PSFLTLSALGAPWAFPPTEE332 pKa = 4.17KK333 pKa = 10.53SVSRR337 pKa = 11.84SSFNHH342 pKa = 6.18HH343 pKa = 6.38SVTGNNDD350 pKa = 3.2PQGRR354 pKa = 11.84RR355 pKa = 11.84WMTFLPKK362 pKa = 8.92PTALQVPNPTGHH374 pKa = 6.93GEE376 pKa = 4.34TPTSSPGPTGPPPTPTEE393 pKa = 3.65IDD395 pKa = 3.48TPIGVLFLAQTSWGDD410 pKa = 4.01FINHH414 pKa = 4.61TWGTKK419 pKa = 9.18PYY421 pKa = 10.64NFGTTSDD428 pKa = 3.96EE429 pKa = 4.56SPNFPPFSRR438 pKa = 11.84DD439 pKa = 3.08TQNRR443 pKa = 11.84PWAVIKK449 pKa = 10.76LKK451 pKa = 11.03SKK453 pKa = 8.85WQLGNKK459 pKa = 7.5QRR461 pKa = 11.84PYY463 pKa = 10.35WYY465 pKa = 7.78DD466 pKa = 3.61TNWWKK471 pKa = 10.48QSRR474 pKa = 11.84TQFGG478 pKa = 3.14
Molecular weight: 55.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
813 |
103 |
478 |
271.0 |
31.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.289 ± 0.59 | 1.107 ± 0.26 |
5.535 ± 1.717 | 3.69 ± 1.154 |
4.92 ± 1.119 | 6.396 ± 0.759 |
2.829 ± 0.412 | 4.182 ± 0.603 |
4.428 ± 0.434 | 6.888 ± 0.795 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.845 ± 0.21 | 4.92 ± 0.709 |
8.118 ± 1.918 | 5.289 ± 0.779 |
9.102 ± 0.442 | 7.503 ± 0.573 |
8.241 ± 0.81 | 3.321 ± 0.711 |
2.706 ± 0.725 | 3.69 ± 0.111 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
