
Brevibacillus brevis (strain 47 / JCM 6285 / NBRC 100599)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Brevibacillus; Brevibacillus brevis
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
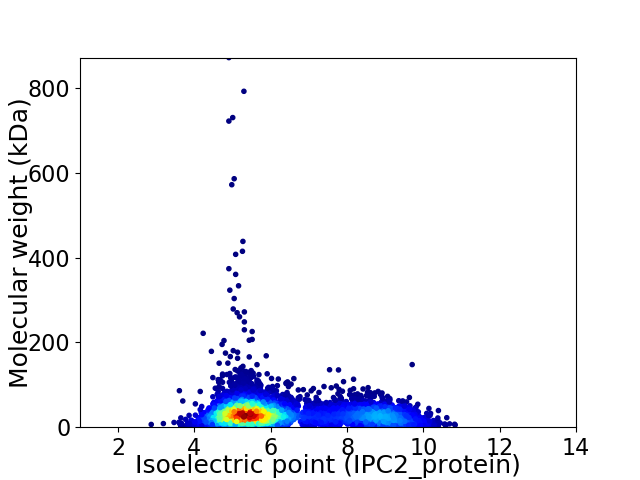
Virtual 2D-PAGE plot for 5887 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0ZKS4|C0ZKS4_BREBN Uncharacterized protein OS=Brevibacillus brevis (strain 47 / JCM 6285 / NBRC 100599) OX=358681 GN=yuzB PE=4 SV=1
MM1 pKa = 7.38FGVKK5 pKa = 9.81HH6 pKa = 5.19FNEE9 pKa = 5.15RR10 pKa = 11.84IGACIPVRR18 pKa = 11.84KK19 pKa = 10.05GEE21 pKa = 4.38SNLACPFFQPAVEE34 pKa = 4.11YY35 pKa = 8.32DD36 pKa = 3.35TGGTSPSSITSADD49 pKa = 3.53FNGDD53 pKa = 3.92SIIDD57 pKa = 3.84LAVVDD62 pKa = 4.37IGSNDD67 pKa = 3.3VSILLNNGSGVFSVSQTIPFNTLFPGSLLGQIAAADD103 pKa = 4.07FNGDD107 pKa = 3.15GHH109 pKa = 7.66IDD111 pKa = 3.9LAVTDD116 pKa = 3.81QTPNDD121 pKa = 3.49RR122 pKa = 11.84VFILLNNGDD131 pKa = 3.99GTFAAPVAYY140 pKa = 8.96STGVFPAVSQSFAIATGDD158 pKa = 3.38FDD160 pKa = 4.37GVNGVDD166 pKa = 3.8LAIANVGQITGTSFITVLLNNGSGVFTMAPGSPFATTPGLAPFDD210 pKa = 3.43IVAADD215 pKa = 3.66FNGDD219 pKa = 3.42GKK221 pKa = 11.33LDD223 pKa = 3.72LATANRR229 pKa = 11.84DD230 pKa = 3.66TNDD233 pKa = 2.58ITIFNGNGDD242 pKa = 3.94GTFQTPGVSFSVLPGGASPLSLVAADD268 pKa = 4.28LNGNGSIDD276 pKa = 3.2IAVANIATSNVTVFLNNGFGSFTEE300 pKa = 4.19AAGSPFSLGTDD311 pKa = 3.81AFPFDD316 pKa = 3.22ITAADD321 pKa = 4.53FNCDD325 pKa = 3.15GAIDD329 pKa = 4.39LATPNAGSDD338 pKa = 3.56NVSVLINNGTGGFSTPFQFPTGAGPFSITVADD370 pKa = 4.11FNGDD374 pKa = 3.12GRR376 pKa = 11.84VDD378 pKa = 2.87IATANEE384 pKa = 3.87GSNNVSVLLNGCVQPPTCPPDD405 pKa = 3.42MVVPCPIVNYY415 pKa = 6.66STPVATCPGVTVTCSPPSGSLFPVGDD441 pKa = 3.77TVVTCTAQDD450 pKa = 2.91IFGNTEE456 pKa = 3.61VCSFTITVIDD466 pKa = 4.25EE467 pKa = 4.78PPTITCPEE475 pKa = 5.46DD476 pKa = 2.89ITVPVLPGRR485 pKa = 11.84TSATVTYY492 pKa = 9.74EE493 pKa = 3.68VTATDD498 pKa = 3.58TCGVTSITCHH508 pKa = 4.81GMTRR512 pKa = 11.84SFSPPAPIAVATFTMDD528 pKa = 3.85FPVGMTMLTCTARR541 pKa = 11.84DD542 pKa = 3.49QSGNTASCSFVVTVTFGVPNSIQPEE567 pKa = 4.31IIRR570 pKa = 11.84VKK572 pKa = 10.41KK573 pKa = 9.85VYY575 pKa = 10.44DD576 pKa = 3.49WVVLQSNFRR585 pKa = 11.84LILDD589 pKa = 4.61LNHH592 pKa = 6.72SRR594 pKa = 11.84DD595 pKa = 3.65DD596 pKa = 3.95QGNQKK601 pKa = 10.52
MM1 pKa = 7.38FGVKK5 pKa = 9.81HH6 pKa = 5.19FNEE9 pKa = 5.15RR10 pKa = 11.84IGACIPVRR18 pKa = 11.84KK19 pKa = 10.05GEE21 pKa = 4.38SNLACPFFQPAVEE34 pKa = 4.11YY35 pKa = 8.32DD36 pKa = 3.35TGGTSPSSITSADD49 pKa = 3.53FNGDD53 pKa = 3.92SIIDD57 pKa = 3.84LAVVDD62 pKa = 4.37IGSNDD67 pKa = 3.3VSILLNNGSGVFSVSQTIPFNTLFPGSLLGQIAAADD103 pKa = 4.07FNGDD107 pKa = 3.15GHH109 pKa = 7.66IDD111 pKa = 3.9LAVTDD116 pKa = 3.81QTPNDD121 pKa = 3.49RR122 pKa = 11.84VFILLNNGDD131 pKa = 3.99GTFAAPVAYY140 pKa = 8.96STGVFPAVSQSFAIATGDD158 pKa = 3.38FDD160 pKa = 4.37GVNGVDD166 pKa = 3.8LAIANVGQITGTSFITVLLNNGSGVFTMAPGSPFATTPGLAPFDD210 pKa = 3.43IVAADD215 pKa = 3.66FNGDD219 pKa = 3.42GKK221 pKa = 11.33LDD223 pKa = 3.72LATANRR229 pKa = 11.84DD230 pKa = 3.66TNDD233 pKa = 2.58ITIFNGNGDD242 pKa = 3.94GTFQTPGVSFSVLPGGASPLSLVAADD268 pKa = 4.28LNGNGSIDD276 pKa = 3.2IAVANIATSNVTVFLNNGFGSFTEE300 pKa = 4.19AAGSPFSLGTDD311 pKa = 3.81AFPFDD316 pKa = 3.22ITAADD321 pKa = 4.53FNCDD325 pKa = 3.15GAIDD329 pKa = 4.39LATPNAGSDD338 pKa = 3.56NVSVLINNGTGGFSTPFQFPTGAGPFSITVADD370 pKa = 4.11FNGDD374 pKa = 3.12GRR376 pKa = 11.84VDD378 pKa = 2.87IATANEE384 pKa = 3.87GSNNVSVLLNGCVQPPTCPPDD405 pKa = 3.42MVVPCPIVNYY415 pKa = 6.66STPVATCPGVTVTCSPPSGSLFPVGDD441 pKa = 3.77TVVTCTAQDD450 pKa = 2.91IFGNTEE456 pKa = 3.61VCSFTITVIDD466 pKa = 4.25EE467 pKa = 4.78PPTITCPEE475 pKa = 5.46DD476 pKa = 2.89ITVPVLPGRR485 pKa = 11.84TSATVTYY492 pKa = 9.74EE493 pKa = 3.68VTATDD498 pKa = 3.58TCGVTSITCHH508 pKa = 4.81GMTRR512 pKa = 11.84SFSPPAPIAVATFTMDD528 pKa = 3.85FPVGMTMLTCTARR541 pKa = 11.84DD542 pKa = 3.49QSGNTASCSFVVTVTFGVPNSIQPEE567 pKa = 4.31IIRR570 pKa = 11.84VKK572 pKa = 10.41KK573 pKa = 9.85VYY575 pKa = 10.44DD576 pKa = 3.49WVVLQSNFRR585 pKa = 11.84LILDD589 pKa = 4.61LNHH592 pKa = 6.72SRR594 pKa = 11.84DD595 pKa = 3.65DD596 pKa = 3.95QGNQKK601 pKa = 10.52
Molecular weight: 61.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0ZAC3|C0ZAC3_BREBN Uncharacterized protein OS=Brevibacillus brevis (strain 47 / JCM 6285 / NBRC 100599) OX=358681 GN=BBR47_17550 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.2PSFNPNNRR10 pKa = 11.84KK11 pKa = 9.37RR12 pKa = 11.84KK13 pKa = 8.62KK14 pKa = 9.81DD15 pKa = 2.98HH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 8.75ILAARR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.84VLSAA44 pKa = 4.11
MM1 pKa = 7.41KK2 pKa = 10.2PSFNPNNRR10 pKa = 11.84KK11 pKa = 9.37RR12 pKa = 11.84KK13 pKa = 8.62KK14 pKa = 9.81DD15 pKa = 2.98HH16 pKa = 6.23GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 8.75ILAARR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.84VLSAA44 pKa = 4.11
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1813859 |
27 |
7745 |
308.1 |
34.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.999 ± 0.031 | 0.765 ± 0.01 |
4.958 ± 0.024 | 7.106 ± 0.039 |
4.132 ± 0.025 | 7.08 ± 0.032 |
2.194 ± 0.016 | 6.647 ± 0.028 |
5.674 ± 0.033 | 10.07 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.902 ± 0.017 | 3.605 ± 0.02 |
4.089 ± 0.024 | 4.296 ± 0.03 |
4.883 ± 0.026 | 5.968 ± 0.026 |
5.641 ± 0.028 | 7.442 ± 0.027 |
1.224 ± 0.014 | 3.323 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
