
Legionella spiritensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales;
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
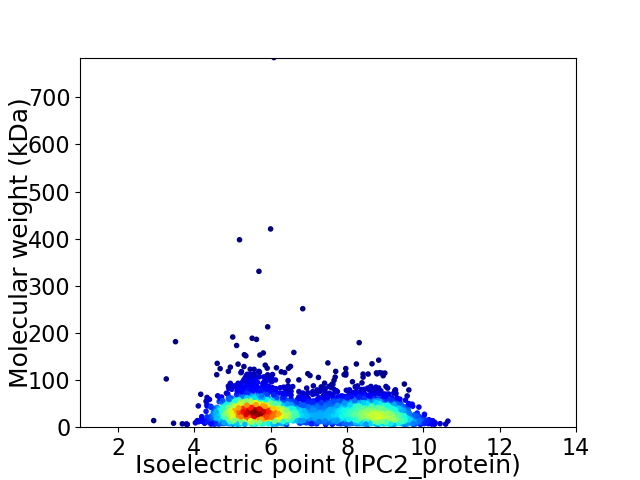
Virtual 2D-PAGE plot for 2908 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
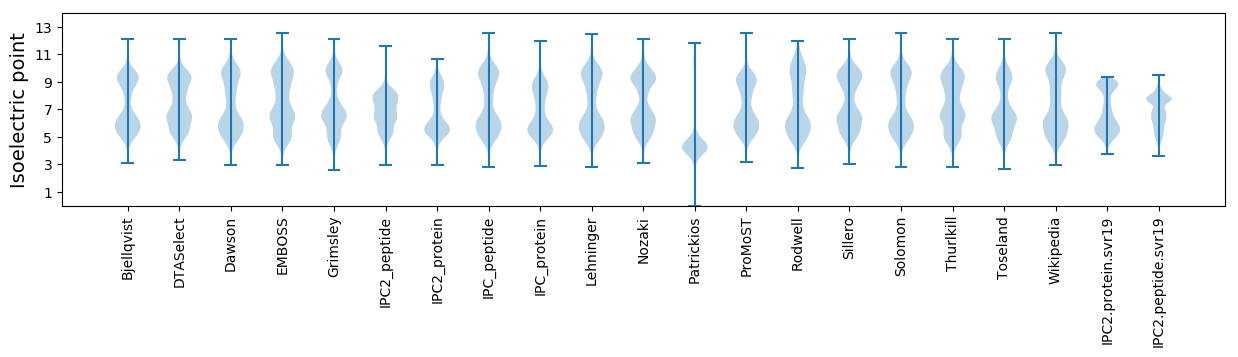
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W0ZAZ1|A0A0W0ZAZ1_LEGSP Uncharacterized protein OS=Legionella spiritensis OX=452 GN=Lspi_0051 PE=4 SV=1
MM1 pKa = 7.47VGVFMVVPSLSGVVQGLEE19 pKa = 4.29GILIKK24 pKa = 9.62NTRR27 pKa = 11.84DD28 pKa = 3.24GGNEE32 pKa = 3.82VAKK35 pKa = 10.7LGTPVHH41 pKa = 6.18QGDD44 pKa = 4.2ILTLLSGNAYY54 pKa = 9.2IHH56 pKa = 5.83MLEE59 pKa = 4.55GFPEE63 pKa = 4.18ALALNRR69 pKa = 11.84PVQIDD74 pKa = 4.75GISPLLKK81 pKa = 10.26AAPQEE86 pKa = 4.31SLVEE90 pKa = 4.1QIVDD94 pKa = 3.26EE95 pKa = 4.59ALAKK99 pKa = 10.91GIDD102 pKa = 3.4PTIILNVLEE111 pKa = 4.03EE112 pKa = 4.15TAAGEE117 pKa = 4.15EE118 pKa = 4.56TIGEE122 pKa = 4.15GGRR125 pKa = 11.84FFVLDD130 pKa = 3.64PLYY133 pKa = 11.09GVGAVTAGYY142 pKa = 6.63PTVGVSVDD150 pKa = 3.7TNSGEE155 pKa = 3.92QHH157 pKa = 5.28VFFYY161 pKa = 10.88EE162 pKa = 4.18PDD164 pKa = 3.52SSEE167 pKa = 3.93QASDD171 pKa = 3.79FVSNPDD177 pKa = 3.32VTEE180 pKa = 4.07TTPFAIGVSSVSVDD194 pKa = 3.42EE195 pKa = 4.49SDD197 pKa = 4.24GLATVVNRR205 pKa = 11.84VQSSTAASEE214 pKa = 4.06ITSVEE219 pKa = 3.98LSTPDD224 pKa = 2.88SNAAVWHH231 pKa = 6.68SGTQTLTATDD241 pKa = 4.78GSWQIMVVRR250 pKa = 11.84SGNNEE255 pKa = 3.08INYY258 pKa = 8.62EE259 pKa = 4.38FKK261 pKa = 10.77QLTPLQHH268 pKa = 7.25PDD270 pKa = 3.05ASDD273 pKa = 3.79PNDD276 pKa = 4.86AITVPITVTATRR288 pKa = 11.84NDD290 pKa = 3.71GASATDD296 pKa = 3.42RR297 pKa = 11.84FSVTIYY303 pKa = 11.04DD304 pKa = 4.43DD305 pKa = 3.97GPKK308 pKa = 9.48IEE310 pKa = 4.26EE311 pKa = 4.39ALVEE315 pKa = 4.63SPLPLNVDD323 pKa = 3.42EE324 pKa = 5.0TNISSGNAVATVNYY338 pKa = 9.54FNTFAGAVHH347 pKa = 6.98FGMDD351 pKa = 4.05DD352 pKa = 3.64TLDD355 pKa = 3.79HH356 pKa = 6.84PGEE359 pKa = 4.32LVFSLGVTADD369 pKa = 3.52QVASGLYY376 pKa = 10.67ANDD379 pKa = 3.72PSAVDD384 pKa = 3.6GKK386 pKa = 10.97GVEE389 pKa = 3.85IQLIEE394 pKa = 4.02EE395 pKa = 4.5GGAIRR400 pKa = 11.84GIAGGIEE407 pKa = 4.44YY408 pKa = 8.03FTISIDD414 pKa = 3.93SNSGEE419 pKa = 3.98MTFAQSEE426 pKa = 4.85AIWHH430 pKa = 6.03GNPANPDD437 pKa = 3.55DD438 pKa = 4.48LASLNTPDD446 pKa = 5.06INDD449 pKa = 3.29LFIRR453 pKa = 11.84VTAIDD458 pKa = 3.81GDD460 pKa = 4.08GDD462 pKa = 3.74KK463 pKa = 11.15ASYY466 pKa = 10.29QLGLGQGIFSINDD479 pKa = 3.74DD480 pKa = 4.26GPGIDD485 pKa = 4.31PGLIVAADD493 pKa = 4.04PLNVDD498 pKa = 3.83EE499 pKa = 5.17TNLATGNAVATANYY513 pKa = 10.24ADD515 pKa = 4.1NFQPAIDD522 pKa = 4.07YY523 pKa = 11.25GEE525 pKa = 5.08DD526 pKa = 3.56GAGQTVYY533 pKa = 10.55EE534 pKa = 4.62LSLNGQPGSGLYY546 pKa = 10.67AIDD549 pKa = 4.2PNQANGQGDD558 pKa = 4.88EE559 pKa = 4.18IQLVQVDD566 pKa = 3.64SHH568 pKa = 5.52TVQGQVNGEE577 pKa = 4.23TYY579 pKa = 9.16FTVTVDD585 pKa = 3.17GGTGEE590 pKa = 4.1VTFAQNRR597 pKa = 11.84AVWHH601 pKa = 6.42SDD603 pKa = 2.81SANANDD609 pKa = 5.23LSTLTTANADD619 pKa = 3.58DD620 pKa = 3.73FTIRR624 pKa = 11.84AIVTDD629 pKa = 3.79ADD631 pKa = 3.88GDD633 pKa = 4.41SVEE636 pKa = 4.11QALNLGQGTFSINDD650 pKa = 3.96DD651 pKa = 3.8GPLVNGNTDD660 pKa = 3.47TAQLVYY666 pKa = 10.84EE667 pKa = 4.66DD668 pKa = 5.24ALPAGNADD676 pKa = 3.55PGQTLNISGSLSSLVDD692 pKa = 4.45FGADD696 pKa = 3.24GPGGFEE702 pKa = 4.21LTTDD706 pKa = 3.76YY707 pKa = 10.4LTTLAAFGLTSDD719 pKa = 4.35GQALSYY725 pKa = 10.45SASEE729 pKa = 4.02NNGIQSVVAKK739 pKa = 10.13DD740 pKa = 3.65YY741 pKa = 11.17LGQDD745 pKa = 3.05VFTFTFDD752 pKa = 3.36KK753 pKa = 9.09TTGNYY758 pKa = 10.55DD759 pKa = 3.19FTLQSHH765 pKa = 7.35LDD767 pKa = 3.7HH768 pKa = 7.11LPPPIAQDD776 pKa = 4.64DD777 pKa = 4.19DD778 pKa = 4.35TQSLPIQLGAIINAIDD794 pKa = 3.85ADD796 pKa = 4.02GDD798 pKa = 4.16KK799 pKa = 11.53VNLGQSFGVHH809 pKa = 5.11VQDD812 pKa = 5.74DD813 pKa = 4.47LPVAQNDD820 pKa = 3.92SVSVNEE826 pKa = 4.85PNAQLNIYY834 pKa = 9.36YY835 pKa = 10.13VVDD838 pKa = 3.46VSGSMGVVDD847 pKa = 4.54PLIGQSRR854 pKa = 11.84LEE856 pKa = 4.07LARR859 pKa = 11.84EE860 pKa = 3.88ALVNLTQEE868 pKa = 4.09YY869 pKa = 10.77ANYY872 pKa = 9.54NGEE875 pKa = 4.15VFVTVVPFASGSGNSGAFATAKK897 pKa = 10.21FDD899 pKa = 3.63ILHH902 pKa = 6.59GEE904 pKa = 4.33LVDD907 pKa = 3.78ALNFIGNLQVSQNPIPGSGTEE928 pKa = 4.2YY929 pKa = 11.25NDD931 pKa = 4.24ALDD934 pKa = 3.75VTQDD938 pKa = 3.61NLEE941 pKa = 4.44LDD943 pKa = 4.73LNPLSQVAGYY953 pKa = 10.31QNVVYY958 pKa = 9.54FLSDD962 pKa = 3.84GVPVPASAGAPAGWQSFVDD981 pKa = 3.71TNNIDD986 pKa = 3.77VIPIGIGDD994 pKa = 4.58FNPGPLEE1001 pKa = 4.05EE1002 pKa = 4.58VGNTGDD1008 pKa = 3.72TVIQVLDD1015 pKa = 3.81PTDD1018 pKa = 3.84LSSTLIEE1025 pKa = 3.97QLDD1028 pKa = 3.81NVVQGNVLDD1037 pKa = 4.08NDD1039 pKa = 3.88AFGGDD1044 pKa = 3.99GPDD1047 pKa = 3.16VPSIVSVSYY1056 pKa = 10.5QGVDD1060 pKa = 2.83HH1061 pKa = 7.73DD1062 pKa = 4.84YY1063 pKa = 11.62DD1064 pKa = 3.78GVNNSVTIIDD1074 pKa = 4.09DD1075 pKa = 3.6FGGRR1079 pKa = 11.84LTFDD1083 pKa = 3.9FTSGDD1088 pKa = 3.53YY1089 pKa = 10.9SYY1091 pKa = 10.83IAPGVDD1097 pKa = 2.93QDD1099 pKa = 3.9EE1100 pKa = 4.41QVQFQYY1106 pKa = 10.68TIRR1109 pKa = 11.84DD1110 pKa = 3.42NDD1112 pKa = 3.74GDD1114 pKa = 4.36LSTANLNIVVNHH1126 pKa = 7.07LDD1128 pKa = 3.85PPSVTLLTKK1137 pKa = 9.74TLEE1140 pKa = 4.21FTRR1143 pKa = 11.84TEE1145 pKa = 4.04QGQQPYY1151 pKa = 10.67GNADD1155 pKa = 3.14VAEE1158 pKa = 4.08YY1159 pKa = 11.29LLLQALGNEE1168 pKa = 4.53SQQVTSVPINGVNLSNTFISVATEE1192 pKa = 3.54PTVNFVSEE1200 pKa = 4.17GAGYY1204 pKa = 10.67RR1205 pKa = 11.84NMVGYY1210 pKa = 7.87YY1211 pKa = 9.95TYY1213 pKa = 11.28DD1214 pKa = 2.92SDD1216 pKa = 5.87GNINDD1221 pKa = 4.91AGFIWVNSSASSSAQNPNGGTNLITNTSGAFGFNQSASFTLPDD1264 pKa = 3.35VSAGEE1269 pKa = 4.01GLGFFLLADD1278 pKa = 4.6GANDD1282 pKa = 3.6SANVNIIRR1290 pKa = 11.84NGNYY1294 pKa = 10.39DD1295 pKa = 4.32SIQSLNNNVSIVNNQVLVGGQALNGSIYY1323 pKa = 9.23FTHH1326 pKa = 7.77DD1327 pKa = 2.56KK1328 pKa = 10.11TLNSDD1333 pKa = 3.51YY1334 pKa = 11.01ATFPEE1339 pKa = 4.45GHH1341 pKa = 7.02ALSGVTFSQDD1351 pKa = 2.99PSLTAYY1357 pKa = 10.39SGLLIIGFEE1366 pKa = 4.23DD1367 pKa = 3.47VHH1369 pKa = 8.99DD1370 pKa = 5.78GGDD1373 pKa = 3.1QDD1375 pKa = 4.22YY1376 pKa = 10.84NDD1378 pKa = 4.28VIFSVDD1384 pKa = 3.49LGANIQNYY1392 pKa = 7.86ATDD1395 pKa = 3.54SSPIIEE1401 pKa = 4.83SVTDD1405 pKa = 3.75PDD1407 pKa = 3.87SAYY1410 pKa = 9.28LTKK1413 pKa = 10.75AVVTTDD1419 pKa = 3.18GFIEE1423 pKa = 5.02GDD1425 pKa = 3.63TLNFLSNANVQFDD1438 pKa = 4.01TVNNQVTVTYY1448 pKa = 10.71NGADD1452 pKa = 2.97HH1453 pKa = 6.34VLNYY1457 pKa = 10.17TLTPDD1462 pKa = 4.02GNDD1465 pKa = 3.13PDD1467 pKa = 5.3FIYY1470 pKa = 10.38TFAAADD1476 pKa = 3.55VNNPVPVGAYY1486 pKa = 9.66EE1487 pKa = 3.98MALKK1491 pKa = 10.37SISFTPVASEE1501 pKa = 3.67ASEE1504 pKa = 4.28GVARR1508 pKa = 11.84DD1509 pKa = 3.58VYY1511 pKa = 10.49VTVTDD1516 pKa = 3.83DD1517 pKa = 3.38TGLSATQATSFNVVINDD1534 pKa = 4.06ILPMDD1539 pKa = 5.22LLAFDD1544 pKa = 4.04QQNEE1548 pKa = 3.91FDD1550 pKa = 4.55FGQGNDD1556 pKa = 3.36TVLIPDD1562 pKa = 3.44QMDD1565 pKa = 3.41GLGQVSGGQGIDD1577 pKa = 3.04TVKK1580 pKa = 9.77FTEE1583 pKa = 3.94EE1584 pKa = 3.95GMNLTAEE1591 pKa = 5.05EE1592 pKa = 4.28IQDD1595 pKa = 3.6HH1596 pKa = 6.9FNGHH1600 pKa = 5.73WEE1602 pKa = 4.14QIDD1605 pKa = 3.6MTEE1608 pKa = 3.84QGSNILNLDD1617 pKa = 3.84LEE1619 pKa = 4.83AVLKK1623 pKa = 10.72LSSGSEE1629 pKa = 4.35LNLQLNNGEE1638 pKa = 4.33NVSVLQIKK1646 pKa = 10.61ADD1648 pKa = 3.87GTDD1651 pKa = 3.6SPDD1654 pKa = 3.44GSVDD1658 pKa = 3.4TVNIDD1663 pKa = 3.49SAHH1666 pKa = 5.14VTDD1669 pKa = 6.13VSDD1672 pKa = 3.67QLPAAGQTAAEE1683 pKa = 4.13QLFKK1687 pKa = 10.52LTNADD1692 pKa = 3.56ASQTAYY1698 pKa = 9.98VHH1700 pKa = 6.04VVSLTQQLPEE1710 pKa = 3.9VHH1712 pKa = 6.09ATSTDD1717 pKa = 3.14GGGGGGGGGG1726 pKa = 3.4
MM1 pKa = 7.47VGVFMVVPSLSGVVQGLEE19 pKa = 4.29GILIKK24 pKa = 9.62NTRR27 pKa = 11.84DD28 pKa = 3.24GGNEE32 pKa = 3.82VAKK35 pKa = 10.7LGTPVHH41 pKa = 6.18QGDD44 pKa = 4.2ILTLLSGNAYY54 pKa = 9.2IHH56 pKa = 5.83MLEE59 pKa = 4.55GFPEE63 pKa = 4.18ALALNRR69 pKa = 11.84PVQIDD74 pKa = 4.75GISPLLKK81 pKa = 10.26AAPQEE86 pKa = 4.31SLVEE90 pKa = 4.1QIVDD94 pKa = 3.26EE95 pKa = 4.59ALAKK99 pKa = 10.91GIDD102 pKa = 3.4PTIILNVLEE111 pKa = 4.03EE112 pKa = 4.15TAAGEE117 pKa = 4.15EE118 pKa = 4.56TIGEE122 pKa = 4.15GGRR125 pKa = 11.84FFVLDD130 pKa = 3.64PLYY133 pKa = 11.09GVGAVTAGYY142 pKa = 6.63PTVGVSVDD150 pKa = 3.7TNSGEE155 pKa = 3.92QHH157 pKa = 5.28VFFYY161 pKa = 10.88EE162 pKa = 4.18PDD164 pKa = 3.52SSEE167 pKa = 3.93QASDD171 pKa = 3.79FVSNPDD177 pKa = 3.32VTEE180 pKa = 4.07TTPFAIGVSSVSVDD194 pKa = 3.42EE195 pKa = 4.49SDD197 pKa = 4.24GLATVVNRR205 pKa = 11.84VQSSTAASEE214 pKa = 4.06ITSVEE219 pKa = 3.98LSTPDD224 pKa = 2.88SNAAVWHH231 pKa = 6.68SGTQTLTATDD241 pKa = 4.78GSWQIMVVRR250 pKa = 11.84SGNNEE255 pKa = 3.08INYY258 pKa = 8.62EE259 pKa = 4.38FKK261 pKa = 10.77QLTPLQHH268 pKa = 7.25PDD270 pKa = 3.05ASDD273 pKa = 3.79PNDD276 pKa = 4.86AITVPITVTATRR288 pKa = 11.84NDD290 pKa = 3.71GASATDD296 pKa = 3.42RR297 pKa = 11.84FSVTIYY303 pKa = 11.04DD304 pKa = 4.43DD305 pKa = 3.97GPKK308 pKa = 9.48IEE310 pKa = 4.26EE311 pKa = 4.39ALVEE315 pKa = 4.63SPLPLNVDD323 pKa = 3.42EE324 pKa = 5.0TNISSGNAVATVNYY338 pKa = 9.54FNTFAGAVHH347 pKa = 6.98FGMDD351 pKa = 4.05DD352 pKa = 3.64TLDD355 pKa = 3.79HH356 pKa = 6.84PGEE359 pKa = 4.32LVFSLGVTADD369 pKa = 3.52QVASGLYY376 pKa = 10.67ANDD379 pKa = 3.72PSAVDD384 pKa = 3.6GKK386 pKa = 10.97GVEE389 pKa = 3.85IQLIEE394 pKa = 4.02EE395 pKa = 4.5GGAIRR400 pKa = 11.84GIAGGIEE407 pKa = 4.44YY408 pKa = 8.03FTISIDD414 pKa = 3.93SNSGEE419 pKa = 3.98MTFAQSEE426 pKa = 4.85AIWHH430 pKa = 6.03GNPANPDD437 pKa = 3.55DD438 pKa = 4.48LASLNTPDD446 pKa = 5.06INDD449 pKa = 3.29LFIRR453 pKa = 11.84VTAIDD458 pKa = 3.81GDD460 pKa = 4.08GDD462 pKa = 3.74KK463 pKa = 11.15ASYY466 pKa = 10.29QLGLGQGIFSINDD479 pKa = 3.74DD480 pKa = 4.26GPGIDD485 pKa = 4.31PGLIVAADD493 pKa = 4.04PLNVDD498 pKa = 3.83EE499 pKa = 5.17TNLATGNAVATANYY513 pKa = 10.24ADD515 pKa = 4.1NFQPAIDD522 pKa = 4.07YY523 pKa = 11.25GEE525 pKa = 5.08DD526 pKa = 3.56GAGQTVYY533 pKa = 10.55EE534 pKa = 4.62LSLNGQPGSGLYY546 pKa = 10.67AIDD549 pKa = 4.2PNQANGQGDD558 pKa = 4.88EE559 pKa = 4.18IQLVQVDD566 pKa = 3.64SHH568 pKa = 5.52TVQGQVNGEE577 pKa = 4.23TYY579 pKa = 9.16FTVTVDD585 pKa = 3.17GGTGEE590 pKa = 4.1VTFAQNRR597 pKa = 11.84AVWHH601 pKa = 6.42SDD603 pKa = 2.81SANANDD609 pKa = 5.23LSTLTTANADD619 pKa = 3.58DD620 pKa = 3.73FTIRR624 pKa = 11.84AIVTDD629 pKa = 3.79ADD631 pKa = 3.88GDD633 pKa = 4.41SVEE636 pKa = 4.11QALNLGQGTFSINDD650 pKa = 3.96DD651 pKa = 3.8GPLVNGNTDD660 pKa = 3.47TAQLVYY666 pKa = 10.84EE667 pKa = 4.66DD668 pKa = 5.24ALPAGNADD676 pKa = 3.55PGQTLNISGSLSSLVDD692 pKa = 4.45FGADD696 pKa = 3.24GPGGFEE702 pKa = 4.21LTTDD706 pKa = 3.76YY707 pKa = 10.4LTTLAAFGLTSDD719 pKa = 4.35GQALSYY725 pKa = 10.45SASEE729 pKa = 4.02NNGIQSVVAKK739 pKa = 10.13DD740 pKa = 3.65YY741 pKa = 11.17LGQDD745 pKa = 3.05VFTFTFDD752 pKa = 3.36KK753 pKa = 9.09TTGNYY758 pKa = 10.55DD759 pKa = 3.19FTLQSHH765 pKa = 7.35LDD767 pKa = 3.7HH768 pKa = 7.11LPPPIAQDD776 pKa = 4.64DD777 pKa = 4.19DD778 pKa = 4.35TQSLPIQLGAIINAIDD794 pKa = 3.85ADD796 pKa = 4.02GDD798 pKa = 4.16KK799 pKa = 11.53VNLGQSFGVHH809 pKa = 5.11VQDD812 pKa = 5.74DD813 pKa = 4.47LPVAQNDD820 pKa = 3.92SVSVNEE826 pKa = 4.85PNAQLNIYY834 pKa = 9.36YY835 pKa = 10.13VVDD838 pKa = 3.46VSGSMGVVDD847 pKa = 4.54PLIGQSRR854 pKa = 11.84LEE856 pKa = 4.07LARR859 pKa = 11.84EE860 pKa = 3.88ALVNLTQEE868 pKa = 4.09YY869 pKa = 10.77ANYY872 pKa = 9.54NGEE875 pKa = 4.15VFVTVVPFASGSGNSGAFATAKK897 pKa = 10.21FDD899 pKa = 3.63ILHH902 pKa = 6.59GEE904 pKa = 4.33LVDD907 pKa = 3.78ALNFIGNLQVSQNPIPGSGTEE928 pKa = 4.2YY929 pKa = 11.25NDD931 pKa = 4.24ALDD934 pKa = 3.75VTQDD938 pKa = 3.61NLEE941 pKa = 4.44LDD943 pKa = 4.73LNPLSQVAGYY953 pKa = 10.31QNVVYY958 pKa = 9.54FLSDD962 pKa = 3.84GVPVPASAGAPAGWQSFVDD981 pKa = 3.71TNNIDD986 pKa = 3.77VIPIGIGDD994 pKa = 4.58FNPGPLEE1001 pKa = 4.05EE1002 pKa = 4.58VGNTGDD1008 pKa = 3.72TVIQVLDD1015 pKa = 3.81PTDD1018 pKa = 3.84LSSTLIEE1025 pKa = 3.97QLDD1028 pKa = 3.81NVVQGNVLDD1037 pKa = 4.08NDD1039 pKa = 3.88AFGGDD1044 pKa = 3.99GPDD1047 pKa = 3.16VPSIVSVSYY1056 pKa = 10.5QGVDD1060 pKa = 2.83HH1061 pKa = 7.73DD1062 pKa = 4.84YY1063 pKa = 11.62DD1064 pKa = 3.78GVNNSVTIIDD1074 pKa = 4.09DD1075 pKa = 3.6FGGRR1079 pKa = 11.84LTFDD1083 pKa = 3.9FTSGDD1088 pKa = 3.53YY1089 pKa = 10.9SYY1091 pKa = 10.83IAPGVDD1097 pKa = 2.93QDD1099 pKa = 3.9EE1100 pKa = 4.41QVQFQYY1106 pKa = 10.68TIRR1109 pKa = 11.84DD1110 pKa = 3.42NDD1112 pKa = 3.74GDD1114 pKa = 4.36LSTANLNIVVNHH1126 pKa = 7.07LDD1128 pKa = 3.85PPSVTLLTKK1137 pKa = 9.74TLEE1140 pKa = 4.21FTRR1143 pKa = 11.84TEE1145 pKa = 4.04QGQQPYY1151 pKa = 10.67GNADD1155 pKa = 3.14VAEE1158 pKa = 4.08YY1159 pKa = 11.29LLLQALGNEE1168 pKa = 4.53SQQVTSVPINGVNLSNTFISVATEE1192 pKa = 3.54PTVNFVSEE1200 pKa = 4.17GAGYY1204 pKa = 10.67RR1205 pKa = 11.84NMVGYY1210 pKa = 7.87YY1211 pKa = 9.95TYY1213 pKa = 11.28DD1214 pKa = 2.92SDD1216 pKa = 5.87GNINDD1221 pKa = 4.91AGFIWVNSSASSSAQNPNGGTNLITNTSGAFGFNQSASFTLPDD1264 pKa = 3.35VSAGEE1269 pKa = 4.01GLGFFLLADD1278 pKa = 4.6GANDD1282 pKa = 3.6SANVNIIRR1290 pKa = 11.84NGNYY1294 pKa = 10.39DD1295 pKa = 4.32SIQSLNNNVSIVNNQVLVGGQALNGSIYY1323 pKa = 9.23FTHH1326 pKa = 7.77DD1327 pKa = 2.56KK1328 pKa = 10.11TLNSDD1333 pKa = 3.51YY1334 pKa = 11.01ATFPEE1339 pKa = 4.45GHH1341 pKa = 7.02ALSGVTFSQDD1351 pKa = 2.99PSLTAYY1357 pKa = 10.39SGLLIIGFEE1366 pKa = 4.23DD1367 pKa = 3.47VHH1369 pKa = 8.99DD1370 pKa = 5.78GGDD1373 pKa = 3.1QDD1375 pKa = 4.22YY1376 pKa = 10.84NDD1378 pKa = 4.28VIFSVDD1384 pKa = 3.49LGANIQNYY1392 pKa = 7.86ATDD1395 pKa = 3.54SSPIIEE1401 pKa = 4.83SVTDD1405 pKa = 3.75PDD1407 pKa = 3.87SAYY1410 pKa = 9.28LTKK1413 pKa = 10.75AVVTTDD1419 pKa = 3.18GFIEE1423 pKa = 5.02GDD1425 pKa = 3.63TLNFLSNANVQFDD1438 pKa = 4.01TVNNQVTVTYY1448 pKa = 10.71NGADD1452 pKa = 2.97HH1453 pKa = 6.34VLNYY1457 pKa = 10.17TLTPDD1462 pKa = 4.02GNDD1465 pKa = 3.13PDD1467 pKa = 5.3FIYY1470 pKa = 10.38TFAAADD1476 pKa = 3.55VNNPVPVGAYY1486 pKa = 9.66EE1487 pKa = 3.98MALKK1491 pKa = 10.37SISFTPVASEE1501 pKa = 3.67ASEE1504 pKa = 4.28GVARR1508 pKa = 11.84DD1509 pKa = 3.58VYY1511 pKa = 10.49VTVTDD1516 pKa = 3.83DD1517 pKa = 3.38TGLSATQATSFNVVINDD1534 pKa = 4.06ILPMDD1539 pKa = 5.22LLAFDD1544 pKa = 4.04QQNEE1548 pKa = 3.91FDD1550 pKa = 4.55FGQGNDD1556 pKa = 3.36TVLIPDD1562 pKa = 3.44QMDD1565 pKa = 3.41GLGQVSGGQGIDD1577 pKa = 3.04TVKK1580 pKa = 9.77FTEE1583 pKa = 3.94EE1584 pKa = 3.95GMNLTAEE1591 pKa = 5.05EE1592 pKa = 4.28IQDD1595 pKa = 3.6HH1596 pKa = 6.9FNGHH1600 pKa = 5.73WEE1602 pKa = 4.14QIDD1605 pKa = 3.6MTEE1608 pKa = 3.84QGSNILNLDD1617 pKa = 3.84LEE1619 pKa = 4.83AVLKK1623 pKa = 10.72LSSGSEE1629 pKa = 4.35LNLQLNNGEE1638 pKa = 4.33NVSVLQIKK1646 pKa = 10.61ADD1648 pKa = 3.87GTDD1651 pKa = 3.6SPDD1654 pKa = 3.44GSVDD1658 pKa = 3.4TVNIDD1663 pKa = 3.49SAHH1666 pKa = 5.14VTDD1669 pKa = 6.13VSDD1672 pKa = 3.67QLPAAGQTAAEE1683 pKa = 4.13QLFKK1687 pKa = 10.52LTNADD1692 pKa = 3.56ASQTAYY1698 pKa = 9.98VHH1700 pKa = 6.04VVSLTQQLPEE1710 pKa = 3.9VHH1712 pKa = 6.09ATSTDD1717 pKa = 3.14GGGGGGGGGG1726 pKa = 3.4
Molecular weight: 181.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W0Z964|A0A0W0Z964_LEGSP UPF0434 protein Lspi_0355 OS=Legionella spiritensis OX=452 GN=lpxK_1 PE=3 SV=1
MM1 pKa = 8.03RR2 pKa = 11.84DD3 pKa = 3.6NVPNWHH9 pKa = 6.99IKK11 pKa = 9.07PAIRR15 pKa = 11.84PQASLRR21 pKa = 11.84ATAFSQKK28 pKa = 10.41NGANYY33 pKa = 9.67PHH35 pKa = 6.67FRR37 pKa = 11.84SAKK40 pKa = 8.93IFVHH44 pKa = 5.51RR45 pKa = 11.84TWKK48 pKa = 10.82AGIFCAAANCLANTKK63 pKa = 10.12SSAVLISCNGGTQACSTRR81 pKa = 11.84ILL83 pKa = 3.65
MM1 pKa = 8.03RR2 pKa = 11.84DD3 pKa = 3.6NVPNWHH9 pKa = 6.99IKK11 pKa = 9.07PAIRR15 pKa = 11.84PQASLRR21 pKa = 11.84ATAFSQKK28 pKa = 10.41NGANYY33 pKa = 9.67PHH35 pKa = 6.67FRR37 pKa = 11.84SAKK40 pKa = 8.93IFVHH44 pKa = 5.51RR45 pKa = 11.84TWKK48 pKa = 10.82AGIFCAAANCLANTKK63 pKa = 10.12SSAVLISCNGGTQACSTRR81 pKa = 11.84ILL83 pKa = 3.65
Molecular weight: 9.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
978053 |
50 |
6909 |
336.3 |
37.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.976 ± 0.041 | 1.204 ± 0.016 |
5.254 ± 0.034 | 5.735 ± 0.045 |
4.372 ± 0.032 | 6.345 ± 0.041 |
2.622 ± 0.024 | 6.938 ± 0.038 |
5.575 ± 0.043 | 10.904 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.524 ± 0.021 | 4.425 ± 0.03 |
4.222 ± 0.025 | 4.573 ± 0.036 |
5.039 ± 0.032 | 6.282 ± 0.035 |
5.243 ± 0.031 | 6.191 ± 0.046 |
1.186 ± 0.018 | 3.388 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
