
Pseudogymnoascus sp. 23342-1-I1
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus; unclassified Pseudogymnoascus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
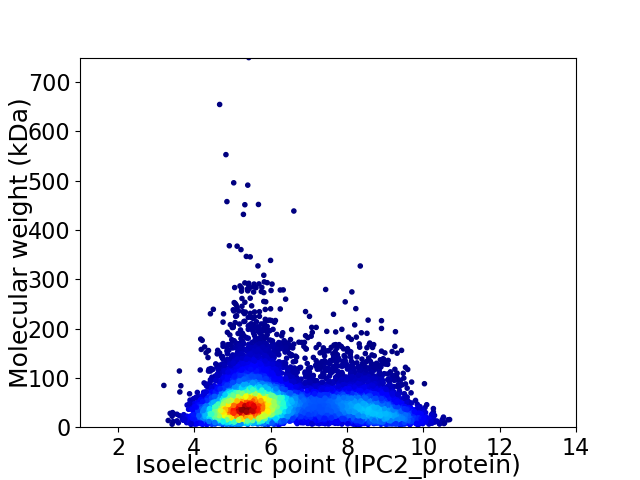
Virtual 2D-PAGE plot for 10680 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
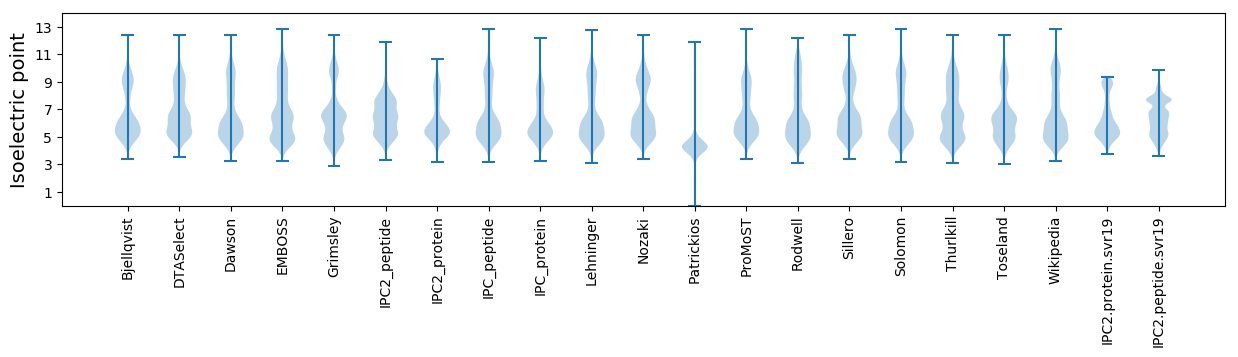
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B8E7C7|A0A1B8E7C7_9PEZI GDP-mannose transporter OS=Pseudogymnoascus sp. 23342-1-I1 OX=1524831 GN=VE03_02658 PE=3 SV=1
MM1 pKa = 7.19QLLNFLPLLLVPAVAAKK18 pKa = 9.04STVYY22 pKa = 10.9LIRR25 pKa = 11.84HH26 pKa = 5.36GEE28 pKa = 3.99KK29 pKa = 10.12PADD32 pKa = 4.0GGDD35 pKa = 3.49GLNAQGLQRR44 pKa = 11.84AQCLRR49 pKa = 11.84NVFGGSSSYY58 pKa = 11.09NIGYY62 pKa = 9.32IMAMTPDD69 pKa = 3.66SDD71 pKa = 3.69GSRR74 pKa = 11.84ARR76 pKa = 11.84PLATVQPLATDD87 pKa = 3.92LGLTVDD93 pKa = 4.21ISCDD97 pKa = 3.54RR98 pKa = 11.84DD99 pKa = 3.73DD100 pKa = 4.24QKK102 pKa = 11.59CVADD106 pKa = 3.81VVNGYY111 pKa = 9.77SGSGNILICWEE122 pKa = 4.27HH123 pKa = 7.23DD124 pKa = 3.36ALTDD128 pKa = 4.04IVDD131 pKa = 3.65EE132 pKa = 5.31LGDD135 pKa = 4.05KK136 pKa = 10.33DD137 pKa = 3.93APSYY141 pKa = 11.0PDD143 pKa = 4.65DD144 pKa = 4.11SFNLIWTDD152 pKa = 3.33PSPYY156 pKa = 10.47SDD158 pKa = 2.72ITATTSEE165 pKa = 4.37NCPGLDD171 pKa = 3.2
MM1 pKa = 7.19QLLNFLPLLLVPAVAAKK18 pKa = 9.04STVYY22 pKa = 10.9LIRR25 pKa = 11.84HH26 pKa = 5.36GEE28 pKa = 3.99KK29 pKa = 10.12PADD32 pKa = 4.0GGDD35 pKa = 3.49GLNAQGLQRR44 pKa = 11.84AQCLRR49 pKa = 11.84NVFGGSSSYY58 pKa = 11.09NIGYY62 pKa = 9.32IMAMTPDD69 pKa = 3.66SDD71 pKa = 3.69GSRR74 pKa = 11.84ARR76 pKa = 11.84PLATVQPLATDD87 pKa = 3.92LGLTVDD93 pKa = 4.21ISCDD97 pKa = 3.54RR98 pKa = 11.84DD99 pKa = 3.73DD100 pKa = 4.24QKK102 pKa = 11.59CVADD106 pKa = 3.81VVNGYY111 pKa = 9.77SGSGNILICWEE122 pKa = 4.27HH123 pKa = 7.23DD124 pKa = 3.36ALTDD128 pKa = 4.04IVDD131 pKa = 3.65EE132 pKa = 5.31LGDD135 pKa = 4.05KK136 pKa = 10.33DD137 pKa = 3.93APSYY141 pKa = 11.0PDD143 pKa = 4.65DD144 pKa = 4.11SFNLIWTDD152 pKa = 3.33PSPYY156 pKa = 10.47SDD158 pKa = 2.72ITATTSEE165 pKa = 4.37NCPGLDD171 pKa = 3.2
Molecular weight: 18.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B8DVZ5|A0A1B8DVZ5_9PEZI WD_REPEATS_REGION domain-containing protein OS=Pseudogymnoascus sp. 23342-1-I1 OX=1524831 GN=VE03_07446 PE=4 SV=1
MM1 pKa = 6.84STSSGPMGVPSAIFGGIPSRR21 pKa = 11.84EE22 pKa = 3.51VDD24 pKa = 3.24IPICAVFIACFVGIGATHH42 pKa = 4.91MTIFQKK48 pKa = 10.29NRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84GHH54 pKa = 5.79KK55 pKa = 9.23FVPNALCFGFSMARR69 pKa = 11.84IATFTMRR76 pKa = 11.84IVWATRR82 pKa = 11.84PTNQNITIAANALVAAGVILLWCINRR108 pKa = 11.84VFATRR113 pKa = 11.84LLGEE117 pKa = 4.01YY118 pKa = 10.07HH119 pKa = 6.58PRR121 pKa = 11.84LYY123 pKa = 10.28TRR125 pKa = 11.84PFFEE129 pKa = 4.14FALRR133 pKa = 11.84RR134 pKa = 11.84LPYY137 pKa = 10.38VVIICCVPMVLVAMVLQIKK156 pKa = 6.18TTNPHH161 pKa = 5.23IRR163 pKa = 11.84VITGILLKK171 pKa = 10.75VVISFFLAFTFSPFIVLAITLLLPGRR197 pKa = 11.84KK198 pKa = 8.85SEE200 pKa = 4.42AEE202 pKa = 4.0VARR205 pKa = 11.84MGKK208 pKa = 10.27GKK210 pKa = 8.92TSTKK214 pKa = 9.98VAIIAIATTLLCWEE228 pKa = 4.54LGIRR232 pKa = 11.84SATMLQKK239 pKa = 10.86LPANAAPWYY248 pKa = 9.29VVASGIEE255 pKa = 4.1LANSMM260 pKa = 4.72
MM1 pKa = 6.84STSSGPMGVPSAIFGGIPSRR21 pKa = 11.84EE22 pKa = 3.51VDD24 pKa = 3.24IPICAVFIACFVGIGATHH42 pKa = 4.91MTIFQKK48 pKa = 10.29NRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84GHH54 pKa = 5.79KK55 pKa = 9.23FVPNALCFGFSMARR69 pKa = 11.84IATFTMRR76 pKa = 11.84IVWATRR82 pKa = 11.84PTNQNITIAANALVAAGVILLWCINRR108 pKa = 11.84VFATRR113 pKa = 11.84LLGEE117 pKa = 4.01YY118 pKa = 10.07HH119 pKa = 6.58PRR121 pKa = 11.84LYY123 pKa = 10.28TRR125 pKa = 11.84PFFEE129 pKa = 4.14FALRR133 pKa = 11.84RR134 pKa = 11.84LPYY137 pKa = 10.38VVIICCVPMVLVAMVLQIKK156 pKa = 6.18TTNPHH161 pKa = 5.23IRR163 pKa = 11.84VITGILLKK171 pKa = 10.75VVISFFLAFTFSPFIVLAITLLLPGRR197 pKa = 11.84KK198 pKa = 8.85SEE200 pKa = 4.42AEE202 pKa = 4.0VARR205 pKa = 11.84MGKK208 pKa = 10.27GKK210 pKa = 8.92TSTKK214 pKa = 9.98VAIIAIATTLLCWEE228 pKa = 4.54LGIRR232 pKa = 11.84SATMLQKK239 pKa = 10.86LPANAAPWYY248 pKa = 9.29VVASGIEE255 pKa = 4.1LANSMM260 pKa = 4.72
Molecular weight: 28.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5193933 |
51 |
6772 |
486.3 |
53.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.96 ± 0.027 | 1.148 ± 0.01 |
5.644 ± 0.017 | 6.4 ± 0.022 |
3.631 ± 0.014 | 7.324 ± 0.021 |
2.2 ± 0.009 | 5.011 ± 0.017 |
4.968 ± 0.023 | 8.776 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.155 ± 0.009 | 3.673 ± 0.012 |
5.963 ± 0.027 | 3.813 ± 0.019 |
5.869 ± 0.021 | 8.07 ± 0.027 |
6.049 ± 0.016 | 6.187 ± 0.016 |
1.411 ± 0.008 | 2.748 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
