
Paenibacillus sp. HB172198
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus; unclassified Paenibacillus
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
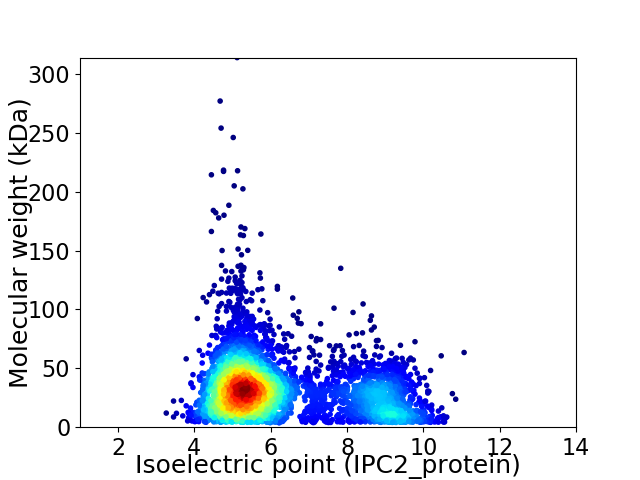
Virtual 2D-PAGE plot for 4162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8XG64|A0A4P8XG64_9BACL Uncharacterized protein OS=Paenibacillus sp. HB172198 OX=2565926 GN=E6C60_0736 PE=4 SV=1
MM1 pKa = 7.42AVINRR6 pKa = 11.84FDD8 pKa = 3.48TTINGAITFTGNTIGLSKK26 pKa = 10.53QLNTNNQGDD35 pKa = 3.75QDD37 pKa = 4.99AIGAMATPNPLSQVNDD53 pKa = 4.47FPPGTTLNFQEE64 pKa = 4.36NAASAVLQLPAQSTVLYY81 pKa = 10.95AEE83 pKa = 5.36LIWSGSYY90 pKa = 10.25NVAGQDD96 pKa = 3.2LSAFINDD103 pKa = 3.13NVSFTTPAGSFSIPFDD119 pKa = 3.67PATAATIFNVLYY131 pKa = 10.61VRR133 pKa = 11.84SADD136 pKa = 3.48VTALVQAAGAGTYY149 pKa = 8.73TVGDD153 pKa = 3.8IVATTSAINNTTNSGGWTLMVAYY176 pKa = 9.25QNPTLPIRR184 pKa = 11.84NMSIFVASEE193 pKa = 4.02FVSTGSGSTPFNISGFFTPTTGPVNGRR220 pKa = 11.84LQVSSLEE227 pKa = 4.15GDD229 pKa = 3.65SNRR232 pKa = 11.84IGDD235 pKa = 3.86QLLFGPTVGTLTPVSGPNNPIDD257 pKa = 3.88NFFASQINGDD267 pKa = 4.81DD268 pKa = 3.8GTLDD272 pKa = 3.27TSGTFGNLNQPLGINADD289 pKa = 3.72GRR291 pKa = 11.84RR292 pKa = 11.84QGYY295 pKa = 9.9DD296 pKa = 2.5ITNIDD301 pKa = 3.8VSPQLVNAQTDD312 pKa = 3.61AVIQGTTSGDD322 pKa = 3.08AYY324 pKa = 10.83VINGVGLQIDD334 pKa = 3.84VNAPRR339 pKa = 11.84FEE341 pKa = 4.23ALKK344 pKa = 10.53SADD347 pKa = 3.45VSVVSPGDD355 pKa = 3.47VITYY359 pKa = 7.64TVEE362 pKa = 3.5ITNTGTADD370 pKa = 3.19ATVVVMFDD378 pKa = 4.02ASPEE382 pKa = 4.23CTTLVPNSVTVDD394 pKa = 3.55GVPQPGATPANLPLGNIIPGQTVIVQYY421 pKa = 10.25QLVVSCVPPSGQYY434 pKa = 10.07VNEE437 pKa = 4.12ALFEE441 pKa = 4.14FQFQSEE447 pKa = 4.42PGGPILTGFGASNILVIQSLQQYY470 pKa = 8.12YY471 pKa = 10.71AINLAAIALAEE482 pKa = 4.28LSQAHH487 pKa = 6.11IMNSEE492 pKa = 4.02AEE494 pKa = 4.12KK495 pKa = 10.63LQTVLGTASLPVITPSLVNLNQLIQVNSSVEE526 pKa = 3.98ATLEE530 pKa = 3.99TLSAQEE536 pKa = 3.67IVLLQRR542 pKa = 11.84FEE544 pKa = 4.76HH545 pKa = 6.66ILRR548 pKa = 11.84GLSVTPGTT556 pKa = 4.21
MM1 pKa = 7.42AVINRR6 pKa = 11.84FDD8 pKa = 3.48TTINGAITFTGNTIGLSKK26 pKa = 10.53QLNTNNQGDD35 pKa = 3.75QDD37 pKa = 4.99AIGAMATPNPLSQVNDD53 pKa = 4.47FPPGTTLNFQEE64 pKa = 4.36NAASAVLQLPAQSTVLYY81 pKa = 10.95AEE83 pKa = 5.36LIWSGSYY90 pKa = 10.25NVAGQDD96 pKa = 3.2LSAFINDD103 pKa = 3.13NVSFTTPAGSFSIPFDD119 pKa = 3.67PATAATIFNVLYY131 pKa = 10.61VRR133 pKa = 11.84SADD136 pKa = 3.48VTALVQAAGAGTYY149 pKa = 8.73TVGDD153 pKa = 3.8IVATTSAINNTTNSGGWTLMVAYY176 pKa = 9.25QNPTLPIRR184 pKa = 11.84NMSIFVASEE193 pKa = 4.02FVSTGSGSTPFNISGFFTPTTGPVNGRR220 pKa = 11.84LQVSSLEE227 pKa = 4.15GDD229 pKa = 3.65SNRR232 pKa = 11.84IGDD235 pKa = 3.86QLLFGPTVGTLTPVSGPNNPIDD257 pKa = 3.88NFFASQINGDD267 pKa = 4.81DD268 pKa = 3.8GTLDD272 pKa = 3.27TSGTFGNLNQPLGINADD289 pKa = 3.72GRR291 pKa = 11.84RR292 pKa = 11.84QGYY295 pKa = 9.9DD296 pKa = 2.5ITNIDD301 pKa = 3.8VSPQLVNAQTDD312 pKa = 3.61AVIQGTTSGDD322 pKa = 3.08AYY324 pKa = 10.83VINGVGLQIDD334 pKa = 3.84VNAPRR339 pKa = 11.84FEE341 pKa = 4.23ALKK344 pKa = 10.53SADD347 pKa = 3.45VSVVSPGDD355 pKa = 3.47VITYY359 pKa = 7.64TVEE362 pKa = 3.5ITNTGTADD370 pKa = 3.19ATVVVMFDD378 pKa = 4.02ASPEE382 pKa = 4.23CTTLVPNSVTVDD394 pKa = 3.55GVPQPGATPANLPLGNIIPGQTVIVQYY421 pKa = 10.25QLVVSCVPPSGQYY434 pKa = 10.07VNEE437 pKa = 4.12ALFEE441 pKa = 4.14FQFQSEE447 pKa = 4.42PGGPILTGFGASNILVIQSLQQYY470 pKa = 8.12YY471 pKa = 10.71AINLAAIALAEE482 pKa = 4.28LSQAHH487 pKa = 6.11IMNSEE492 pKa = 4.02AEE494 pKa = 4.12KK495 pKa = 10.63LQTVLGTASLPVITPSLVNLNQLIQVNSSVEE526 pKa = 3.98ATLEE530 pKa = 3.99TLSAQEE536 pKa = 3.67IVLLQRR542 pKa = 11.84FEE544 pKa = 4.76HH545 pKa = 6.66ILRR548 pKa = 11.84GLSVTPGTT556 pKa = 4.21
Molecular weight: 58.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8XHF3|A0A4P8XHF3_9BACL ATP-dependent Clp protease proteolytic subunit OS=Paenibacillus sp. HB172198 OX=2565926 GN=clpP PE=3 SV=1
MM1 pKa = 7.27GASSSVFRR9 pKa = 11.84VRR11 pKa = 11.84SALNRR16 pKa = 11.84AARR19 pKa = 11.84TSSSVFGVRR28 pKa = 11.84WARR31 pKa = 11.84GRR33 pKa = 11.84TARR36 pKa = 11.84TSSSVFRR43 pKa = 11.84VRR45 pKa = 11.84WALNRR50 pKa = 11.84AARR53 pKa = 11.84VSSSVFRR60 pKa = 11.84TRR62 pKa = 11.84SARR65 pKa = 11.84KK66 pKa = 8.5RR67 pKa = 11.84AAHH70 pKa = 6.32ASSSVFRR77 pKa = 11.84VHH79 pKa = 7.21LLLTRR84 pKa = 11.84RR85 pKa = 11.84CGSSSSVFDD94 pKa = 4.06VRR96 pKa = 11.84WALNRR101 pKa = 11.84AARR104 pKa = 11.84VSSSVFGVQLLQTRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84GSSSSVFDD128 pKa = 3.22VHH130 pKa = 6.51WALNRR135 pKa = 11.84AASASSSVFGVHH147 pKa = 5.23WALNRR152 pKa = 11.84TARR155 pKa = 11.84VSSSVFGVHH164 pKa = 5.54WARR167 pKa = 11.84GRR169 pKa = 11.84AAGASSSVFRR179 pKa = 11.84VHH181 pKa = 7.39LLQTRR186 pKa = 11.84RR187 pKa = 11.84CGSSSSVFDD196 pKa = 4.06VRR198 pKa = 11.84WALNRR203 pKa = 11.84AARR206 pKa = 11.84VSSSVFRR213 pKa = 11.84VRR215 pKa = 11.84WALNRR220 pKa = 11.84AAGASSSVFRR230 pKa = 11.84VQLLQTCRR238 pKa = 11.84RR239 pKa = 11.84GSSSSVFDD247 pKa = 4.06VRR249 pKa = 11.84WALNRR254 pKa = 11.84VARR257 pKa = 11.84VSSSVFRR264 pKa = 11.84VRR266 pKa = 11.84WARR269 pKa = 11.84GRR271 pKa = 11.84ATGASSSVFRR281 pKa = 11.84TRR283 pKa = 11.84STRR286 pKa = 11.84KK287 pKa = 8.02RR288 pKa = 11.84AARR291 pKa = 11.84ASSSVFRR298 pKa = 11.84TRR300 pKa = 11.84SPRR303 pKa = 11.84KK304 pKa = 8.77RR305 pKa = 11.84ATHH308 pKa = 6.29ASSSVFRR315 pKa = 11.84VRR317 pKa = 11.84WARR320 pKa = 11.84GRR322 pKa = 11.84AAGASSSVFRR332 pKa = 11.84VRR334 pKa = 11.84WARR337 pKa = 11.84GRR339 pKa = 11.84AARR342 pKa = 11.84ASSSVFGVHH351 pKa = 5.95WARR354 pKa = 11.84GPAAGASSSVFRR366 pKa = 11.84TRR368 pKa = 11.84STRR371 pKa = 11.84KK372 pKa = 7.56QAARR376 pKa = 11.84ASSSVFRR383 pKa = 11.84VRR385 pKa = 11.84SARR388 pKa = 11.84KK389 pKa = 8.77RR390 pKa = 11.84DD391 pKa = 3.37AGTSSSVFRR400 pKa = 11.84VHH402 pKa = 7.39LLQTRR407 pKa = 11.84RR408 pKa = 11.84CGSSSSVFDD417 pKa = 4.06VRR419 pKa = 11.84WALNRR424 pKa = 11.84TARR427 pKa = 11.84VSSSVFRR434 pKa = 11.84TRR436 pKa = 11.84STRR439 pKa = 11.84KK440 pKa = 8.02RR441 pKa = 11.84AARR444 pKa = 11.84ASSSVFRR451 pKa = 11.84VRR453 pKa = 11.84SARR456 pKa = 11.84KK457 pKa = 8.77RR458 pKa = 11.84DD459 pKa = 3.37AGTSSSVFRR468 pKa = 11.84LRR470 pKa = 11.84WALNRR475 pKa = 11.84TAHH478 pKa = 6.03TSSSVFRR485 pKa = 11.84TRR487 pKa = 11.84WARR490 pKa = 11.84GRR492 pKa = 11.84AARR495 pKa = 11.84ASSSVFRR502 pKa = 11.84TRR504 pKa = 11.84STRR507 pKa = 11.84KK508 pKa = 7.44RR509 pKa = 11.84TARR512 pKa = 11.84ASNSVFRR519 pKa = 11.84TRR521 pKa = 11.84SARR524 pKa = 11.84KK525 pKa = 7.85RR526 pKa = 11.84AARR529 pKa = 11.84ASSSVFRR536 pKa = 11.84TRR538 pKa = 11.84SPRR541 pKa = 11.84SRR543 pKa = 11.84IPSPTQKK550 pKa = 8.81ITPSPYY556 pKa = 8.79FRR558 pKa = 11.84PRR560 pKa = 11.84VILFTAYY567 pKa = 10.79VLDD570 pKa = 4.04EE571 pKa = 4.28TT572 pKa = 5.17
MM1 pKa = 7.27GASSSVFRR9 pKa = 11.84VRR11 pKa = 11.84SALNRR16 pKa = 11.84AARR19 pKa = 11.84TSSSVFGVRR28 pKa = 11.84WARR31 pKa = 11.84GRR33 pKa = 11.84TARR36 pKa = 11.84TSSSVFRR43 pKa = 11.84VRR45 pKa = 11.84WALNRR50 pKa = 11.84AARR53 pKa = 11.84VSSSVFRR60 pKa = 11.84TRR62 pKa = 11.84SARR65 pKa = 11.84KK66 pKa = 8.5RR67 pKa = 11.84AAHH70 pKa = 6.32ASSSVFRR77 pKa = 11.84VHH79 pKa = 7.21LLLTRR84 pKa = 11.84RR85 pKa = 11.84CGSSSSVFDD94 pKa = 4.06VRR96 pKa = 11.84WALNRR101 pKa = 11.84AARR104 pKa = 11.84VSSSVFGVQLLQTRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84GSSSSVFDD128 pKa = 3.22VHH130 pKa = 6.51WALNRR135 pKa = 11.84AASASSSVFGVHH147 pKa = 5.23WALNRR152 pKa = 11.84TARR155 pKa = 11.84VSSSVFGVHH164 pKa = 5.54WARR167 pKa = 11.84GRR169 pKa = 11.84AAGASSSVFRR179 pKa = 11.84VHH181 pKa = 7.39LLQTRR186 pKa = 11.84RR187 pKa = 11.84CGSSSSVFDD196 pKa = 4.06VRR198 pKa = 11.84WALNRR203 pKa = 11.84AARR206 pKa = 11.84VSSSVFRR213 pKa = 11.84VRR215 pKa = 11.84WALNRR220 pKa = 11.84AAGASSSVFRR230 pKa = 11.84VQLLQTCRR238 pKa = 11.84RR239 pKa = 11.84GSSSSVFDD247 pKa = 4.06VRR249 pKa = 11.84WALNRR254 pKa = 11.84VARR257 pKa = 11.84VSSSVFRR264 pKa = 11.84VRR266 pKa = 11.84WARR269 pKa = 11.84GRR271 pKa = 11.84ATGASSSVFRR281 pKa = 11.84TRR283 pKa = 11.84STRR286 pKa = 11.84KK287 pKa = 8.02RR288 pKa = 11.84AARR291 pKa = 11.84ASSSVFRR298 pKa = 11.84TRR300 pKa = 11.84SPRR303 pKa = 11.84KK304 pKa = 8.77RR305 pKa = 11.84ATHH308 pKa = 6.29ASSSVFRR315 pKa = 11.84VRR317 pKa = 11.84WARR320 pKa = 11.84GRR322 pKa = 11.84AAGASSSVFRR332 pKa = 11.84VRR334 pKa = 11.84WARR337 pKa = 11.84GRR339 pKa = 11.84AARR342 pKa = 11.84ASSSVFGVHH351 pKa = 5.95WARR354 pKa = 11.84GPAAGASSSVFRR366 pKa = 11.84TRR368 pKa = 11.84STRR371 pKa = 11.84KK372 pKa = 7.56QAARR376 pKa = 11.84ASSSVFRR383 pKa = 11.84VRR385 pKa = 11.84SARR388 pKa = 11.84KK389 pKa = 8.77RR390 pKa = 11.84DD391 pKa = 3.37AGTSSSVFRR400 pKa = 11.84VHH402 pKa = 7.39LLQTRR407 pKa = 11.84RR408 pKa = 11.84CGSSSSVFDD417 pKa = 4.06VRR419 pKa = 11.84WALNRR424 pKa = 11.84TARR427 pKa = 11.84VSSSVFRR434 pKa = 11.84TRR436 pKa = 11.84STRR439 pKa = 11.84KK440 pKa = 8.02RR441 pKa = 11.84AARR444 pKa = 11.84ASSSVFRR451 pKa = 11.84VRR453 pKa = 11.84SARR456 pKa = 11.84KK457 pKa = 8.77RR458 pKa = 11.84DD459 pKa = 3.37AGTSSSVFRR468 pKa = 11.84LRR470 pKa = 11.84WALNRR475 pKa = 11.84TAHH478 pKa = 6.03TSSSVFRR485 pKa = 11.84TRR487 pKa = 11.84WARR490 pKa = 11.84GRR492 pKa = 11.84AARR495 pKa = 11.84ASSSVFRR502 pKa = 11.84TRR504 pKa = 11.84STRR507 pKa = 11.84KK508 pKa = 7.44RR509 pKa = 11.84TARR512 pKa = 11.84ASNSVFRR519 pKa = 11.84TRR521 pKa = 11.84SARR524 pKa = 11.84KK525 pKa = 7.85RR526 pKa = 11.84AARR529 pKa = 11.84ASSSVFRR536 pKa = 11.84TRR538 pKa = 11.84SPRR541 pKa = 11.84SRR543 pKa = 11.84IPSPTQKK550 pKa = 8.81ITPSPYY556 pKa = 8.79FRR558 pKa = 11.84PRR560 pKa = 11.84VILFTAYY567 pKa = 10.79VLDD570 pKa = 4.04EE571 pKa = 4.28TT572 pKa = 5.17
Molecular weight: 63.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276428 |
37 |
2836 |
306.7 |
34.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.23 ± 0.046 | 0.778 ± 0.011 |
5.072 ± 0.026 | 7.203 ± 0.045 |
3.865 ± 0.027 | 7.455 ± 0.037 |
2.267 ± 0.021 | 6.203 ± 0.033 |
5.055 ± 0.036 | 10.189 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.983 ± 0.02 | 3.492 ± 0.028 |
4.22 ± 0.025 | 4.366 ± 0.03 |
5.25 ± 0.035 | 6.596 ± 0.031 |
5.109 ± 0.026 | 6.962 ± 0.03 |
1.257 ± 0.016 | 3.446 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
