
Mesotoga prima MesG1.Ag.4.2
Taxonomy: cellular organisms; Bacteria; Thermotogae; Thermotogae; Kosmotogales; Kosmotogaceae; Mesotoga; Mesotoga prima
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
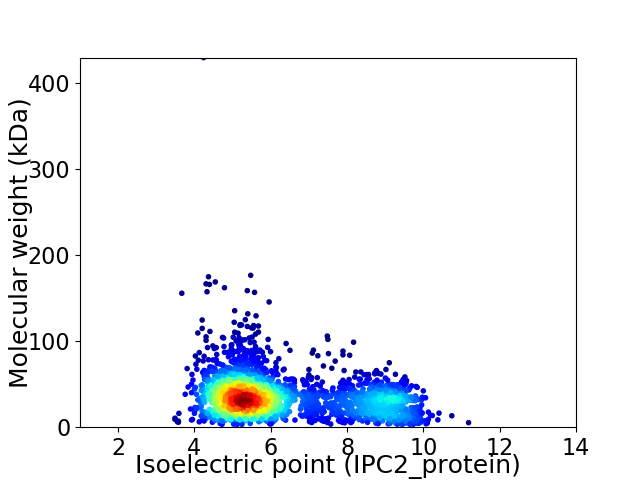
Virtual 2D-PAGE plot for 2499 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I2F6S8|I2F6S8_9BACT UPF0316 protein Theba_1988 OS=Mesotoga prima MesG1.Ag.4.2 OX=660470 GN=Theba_1988 PE=3 SV=1
MM1 pKa = 7.72KK2 pKa = 9.01KK3 pKa = 8.3TLLITLLLGFMLFVSGCFKK22 pKa = 10.77INNPPVISLTGSVSEE37 pKa = 4.33GQEE40 pKa = 3.89IAPMTLVTFEE50 pKa = 4.17WEE52 pKa = 3.72ITDD55 pKa = 4.43FDD57 pKa = 4.1DD58 pKa = 4.82TEE60 pKa = 4.33VTSEE64 pKa = 3.89FKK66 pKa = 10.74LYY68 pKa = 10.79RR69 pKa = 11.84NGAEE73 pKa = 3.98VLSVVDD79 pKa = 3.7TGEE82 pKa = 3.55AMYY85 pKa = 11.29ALDD88 pKa = 4.45EE89 pKa = 4.45EE90 pKa = 4.86GKK92 pKa = 10.33YY93 pKa = 9.46VAEE96 pKa = 4.38VVATDD101 pKa = 4.12SMGAQSKK108 pKa = 7.36EE109 pKa = 4.27TISFKK114 pKa = 10.23ATNLKK119 pKa = 10.47RR120 pKa = 11.84EE121 pKa = 4.4LFQSGAGLMYY131 pKa = 10.17FDD133 pKa = 4.33YY134 pKa = 11.13SDD136 pKa = 3.58YY137 pKa = 10.76FQYY140 pKa = 10.6IEE142 pKa = 5.19DD143 pKa = 3.73EE144 pKa = 4.17AFGIRR149 pKa = 11.84FYY151 pKa = 10.33KK152 pKa = 9.15TASEE156 pKa = 4.46EE157 pKa = 3.84ILPAEE162 pKa = 4.23EE163 pKa = 3.78VTVRR167 pKa = 11.84KK168 pKa = 9.71RR169 pKa = 11.84LNLLDD174 pKa = 3.98TDD176 pKa = 4.48DD177 pKa = 5.69DD178 pKa = 4.31GMYY181 pKa = 8.79DD182 pKa = 3.13TWEE185 pKa = 4.78EE186 pKa = 4.18IPNDD190 pKa = 3.9RR191 pKa = 11.84PDD193 pKa = 3.46LQTYY197 pKa = 9.79VYY199 pKa = 10.54LPGYY203 pKa = 9.92LIDD206 pKa = 5.8YY207 pKa = 7.71DD208 pKa = 4.47TLWVPYY214 pKa = 9.74GFEE217 pKa = 4.09YY218 pKa = 11.07GEE220 pKa = 4.02KK221 pKa = 10.22SIRR224 pKa = 11.84AYY226 pKa = 10.02YY227 pKa = 10.52AGVEE231 pKa = 4.45GPYY234 pKa = 10.03SGYY237 pKa = 10.54VLFYY241 pKa = 10.75KK242 pKa = 10.48FAEE245 pKa = 4.31VGFDD249 pKa = 3.72LNTVYY254 pKa = 10.85ARR256 pKa = 11.84WVEE259 pKa = 3.6AHH261 pKa = 6.19VFGNTLNDD269 pKa = 3.16YY270 pKa = 11.1GEE272 pKa = 4.41AYY274 pKa = 9.74KK275 pKa = 10.95AFALRR280 pKa = 11.84EE281 pKa = 4.06RR282 pKa = 11.84YY283 pKa = 10.11DD284 pKa = 3.48SDD286 pKa = 3.43EE287 pKa = 4.46KK288 pKa = 11.13PIISLSTPDD297 pKa = 4.02GDD299 pKa = 4.36IGPGDD304 pKa = 4.02SFTVSVLTEE313 pKa = 4.13NVASFADD320 pKa = 4.49LYY322 pKa = 10.82DD323 pKa = 3.28VRR325 pKa = 11.84YY326 pKa = 9.73VQLSLWNSEE335 pKa = 3.84GLVLEE340 pKa = 4.53GVEE343 pKa = 4.98FGNFMAGLKK352 pKa = 10.47DD353 pKa = 3.3VGTFKK358 pKa = 11.1SSDD361 pKa = 3.55EE362 pKa = 3.97AVTLYY367 pKa = 10.8KK368 pKa = 10.83AFLNGVDD375 pKa = 3.66EE376 pKa = 5.79AEE378 pKa = 4.33AADD381 pKa = 4.98DD382 pKa = 4.03VFATLTFSVPEE393 pKa = 4.22DD394 pKa = 3.81FEE396 pKa = 5.93GEE398 pKa = 4.24LIAVGLAYY406 pKa = 9.94EE407 pKa = 5.37GYY409 pKa = 7.99WDD411 pKa = 3.81TYY413 pKa = 8.54GTYY416 pKa = 10.12PDD418 pKa = 4.52LPNPAFRR425 pKa = 11.84DD426 pKa = 3.41VDD428 pKa = 3.66NGSVDD433 pKa = 3.57GFIVDD438 pKa = 3.95HH439 pKa = 6.41EE440 pKa = 4.76PIVFLEE446 pKa = 4.08
MM1 pKa = 7.72KK2 pKa = 9.01KK3 pKa = 8.3TLLITLLLGFMLFVSGCFKK22 pKa = 10.77INNPPVISLTGSVSEE37 pKa = 4.33GQEE40 pKa = 3.89IAPMTLVTFEE50 pKa = 4.17WEE52 pKa = 3.72ITDD55 pKa = 4.43FDD57 pKa = 4.1DD58 pKa = 4.82TEE60 pKa = 4.33VTSEE64 pKa = 3.89FKK66 pKa = 10.74LYY68 pKa = 10.79RR69 pKa = 11.84NGAEE73 pKa = 3.98VLSVVDD79 pKa = 3.7TGEE82 pKa = 3.55AMYY85 pKa = 11.29ALDD88 pKa = 4.45EE89 pKa = 4.45EE90 pKa = 4.86GKK92 pKa = 10.33YY93 pKa = 9.46VAEE96 pKa = 4.38VVATDD101 pKa = 4.12SMGAQSKK108 pKa = 7.36EE109 pKa = 4.27TISFKK114 pKa = 10.23ATNLKK119 pKa = 10.47RR120 pKa = 11.84EE121 pKa = 4.4LFQSGAGLMYY131 pKa = 10.17FDD133 pKa = 4.33YY134 pKa = 11.13SDD136 pKa = 3.58YY137 pKa = 10.76FQYY140 pKa = 10.6IEE142 pKa = 5.19DD143 pKa = 3.73EE144 pKa = 4.17AFGIRR149 pKa = 11.84FYY151 pKa = 10.33KK152 pKa = 9.15TASEE156 pKa = 4.46EE157 pKa = 3.84ILPAEE162 pKa = 4.23EE163 pKa = 3.78VTVRR167 pKa = 11.84KK168 pKa = 9.71RR169 pKa = 11.84LNLLDD174 pKa = 3.98TDD176 pKa = 4.48DD177 pKa = 5.69DD178 pKa = 4.31GMYY181 pKa = 8.79DD182 pKa = 3.13TWEE185 pKa = 4.78EE186 pKa = 4.18IPNDD190 pKa = 3.9RR191 pKa = 11.84PDD193 pKa = 3.46LQTYY197 pKa = 9.79VYY199 pKa = 10.54LPGYY203 pKa = 9.92LIDD206 pKa = 5.8YY207 pKa = 7.71DD208 pKa = 4.47TLWVPYY214 pKa = 9.74GFEE217 pKa = 4.09YY218 pKa = 11.07GEE220 pKa = 4.02KK221 pKa = 10.22SIRR224 pKa = 11.84AYY226 pKa = 10.02YY227 pKa = 10.52AGVEE231 pKa = 4.45GPYY234 pKa = 10.03SGYY237 pKa = 10.54VLFYY241 pKa = 10.75KK242 pKa = 10.48FAEE245 pKa = 4.31VGFDD249 pKa = 3.72LNTVYY254 pKa = 10.85ARR256 pKa = 11.84WVEE259 pKa = 3.6AHH261 pKa = 6.19VFGNTLNDD269 pKa = 3.16YY270 pKa = 11.1GEE272 pKa = 4.41AYY274 pKa = 9.74KK275 pKa = 10.95AFALRR280 pKa = 11.84EE281 pKa = 4.06RR282 pKa = 11.84YY283 pKa = 10.11DD284 pKa = 3.48SDD286 pKa = 3.43EE287 pKa = 4.46KK288 pKa = 11.13PIISLSTPDD297 pKa = 4.02GDD299 pKa = 4.36IGPGDD304 pKa = 4.02SFTVSVLTEE313 pKa = 4.13NVASFADD320 pKa = 4.49LYY322 pKa = 10.82DD323 pKa = 3.28VRR325 pKa = 11.84YY326 pKa = 9.73VQLSLWNSEE335 pKa = 3.84GLVLEE340 pKa = 4.53GVEE343 pKa = 4.98FGNFMAGLKK352 pKa = 10.47DD353 pKa = 3.3VGTFKK358 pKa = 11.1SSDD361 pKa = 3.55EE362 pKa = 3.97AVTLYY367 pKa = 10.8KK368 pKa = 10.83AFLNGVDD375 pKa = 3.66EE376 pKa = 5.79AEE378 pKa = 4.33AADD381 pKa = 4.98DD382 pKa = 4.03VFATLTFSVPEE393 pKa = 4.22DD394 pKa = 3.81FEE396 pKa = 5.93GEE398 pKa = 4.24LIAVGLAYY406 pKa = 9.94EE407 pKa = 5.37GYY409 pKa = 7.99WDD411 pKa = 3.81TYY413 pKa = 8.54GTYY416 pKa = 10.12PDD418 pKa = 4.52LPNPAFRR425 pKa = 11.84DD426 pKa = 3.41VDD428 pKa = 3.66NGSVDD433 pKa = 3.57GFIVDD438 pKa = 3.95HH439 pKa = 6.41EE440 pKa = 4.76PIVFLEE446 pKa = 4.08
Molecular weight: 50.1 kDa
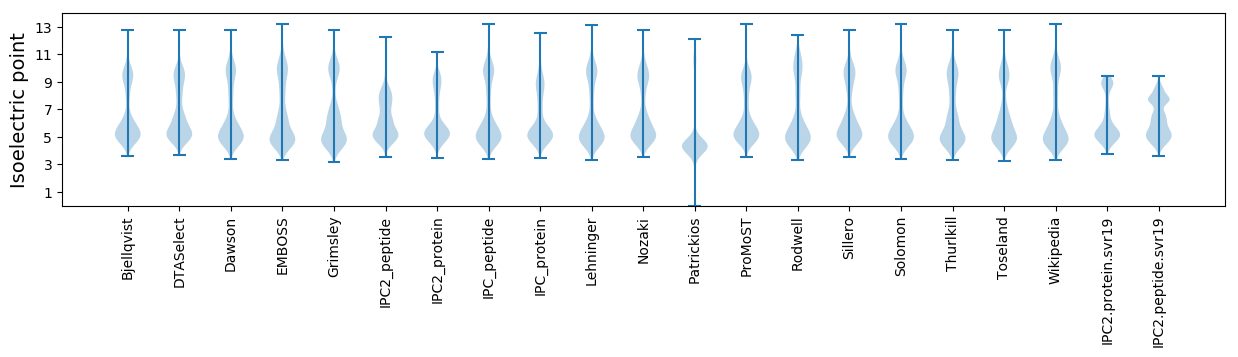
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I2F3P3|I2F3P3_9BACT Na+-transporting NADH:ubiquinone oxidoreductase subunit NqrE OS=Mesotoga prima MesG1.Ag.4.2 OX=660470 GN=Theba_0832 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.16QPSRR9 pKa = 11.84VKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84THH16 pKa = 6.49GFLVRR21 pKa = 11.84SRR23 pKa = 11.84TVGGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.61GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.16QPSRR9 pKa = 11.84VKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84THH16 pKa = 6.49GFLVRR21 pKa = 11.84SRR23 pKa = 11.84TVGGRR28 pKa = 11.84RR29 pKa = 11.84VLASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.61GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
811598 |
29 |
3898 |
324.8 |
36.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.618 ± 0.041 | 0.792 ± 0.019 |
5.404 ± 0.033 | 7.681 ± 0.06 |
5.019 ± 0.039 | 7.404 ± 0.04 |
1.469 ± 0.018 | 7.618 ± 0.042 |
5.951 ± 0.05 | 9.999 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.637 ± 0.023 | 3.783 ± 0.025 |
3.757 ± 0.028 | 2.218 ± 0.018 |
5.379 ± 0.045 | 7.444 ± 0.046 |
4.849 ± 0.032 | 7.516 ± 0.039 |
1.051 ± 0.019 | 3.412 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
