
Avon-Heathcote Estuary associated circular virus 20
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.34
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
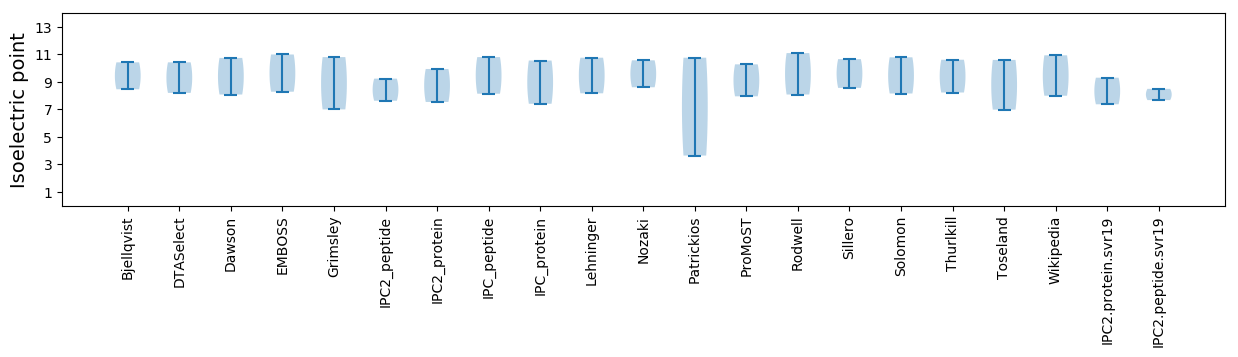
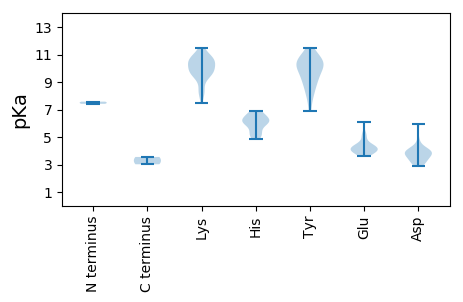
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0C5IMJ6|A0A0C5IMJ6_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 20 OX=1618244 PE=4 SV=1
MM1 pKa = 7.42SRR3 pKa = 11.84IRR5 pKa = 11.84NMMVTIYY12 pKa = 10.66FEE14 pKa = 6.12DD15 pKa = 3.93NGCPDD20 pKa = 3.81PPEE23 pKa = 4.35IRR25 pKa = 11.84AMLGALPNLRR35 pKa = 11.84YY36 pKa = 10.37YY37 pKa = 9.77IGQMEE42 pKa = 4.61KK43 pKa = 10.58CPSTGRR49 pKa = 11.84FHH51 pKa = 6.11FQGYY55 pKa = 9.81IEE57 pKa = 5.3FDD59 pKa = 2.9SAMYY63 pKa = 9.63VKK65 pKa = 10.47KK66 pKa = 10.73ISAQLNNCHH75 pKa = 6.17IEE77 pKa = 4.0ARR79 pKa = 11.84KK80 pKa = 8.34GTQLQAIAYY89 pKa = 6.89CKK91 pKa = 9.78KK92 pKa = 9.9HH93 pKa = 4.86EE94 pKa = 4.23TRR96 pKa = 11.84VGNWIEE102 pKa = 3.73YY103 pKa = 10.0GEE105 pKa = 4.24RR106 pKa = 11.84AKK108 pKa = 10.87QGTRR112 pKa = 11.84SDD114 pKa = 3.88LDD116 pKa = 3.94DD117 pKa = 3.8VAEE120 pKa = 4.82CIYY123 pKa = 10.42GGKK126 pKa = 9.72SIRR129 pKa = 11.84EE130 pKa = 3.94IQEE133 pKa = 3.67EE134 pKa = 4.33HH135 pKa = 6.23PRR137 pKa = 11.84AYY139 pKa = 10.25MMYY142 pKa = 10.26SKK144 pKa = 10.62GIKK147 pKa = 9.62DD148 pKa = 4.06LYY150 pKa = 8.66YY151 pKa = 9.6TVNNIEE157 pKa = 4.45PRR159 pKa = 11.84SEE161 pKa = 4.11PPVVIYY167 pKa = 10.35CYY169 pKa = 10.62GQTGTGKK176 pKa = 9.6SRR178 pKa = 11.84YY179 pKa = 8.9AKK181 pKa = 9.73RR182 pKa = 11.84YY183 pKa = 8.23SDD185 pKa = 3.43NVYY188 pKa = 10.13FKK190 pKa = 11.5NMSCKK195 pKa = 9.16WWDD198 pKa = 3.72GYY200 pKa = 11.35NGQDD204 pKa = 3.02VVVFDD209 pKa = 5.97DD210 pKa = 4.16IRR212 pKa = 11.84GDD214 pKa = 3.4TFKK217 pKa = 11.07YY218 pKa = 10.44HH219 pKa = 6.1EE220 pKa = 5.06LLRR223 pKa = 11.84ILDD226 pKa = 3.37RR227 pKa = 11.84YY228 pKa = 9.12VRR230 pKa = 11.84HH231 pKa = 6.11EE232 pKa = 4.47CDD234 pKa = 3.26SPNAPMVGIPCMLNIRR250 pKa = 11.84GAASTSIRR258 pKa = 11.84LL259 pKa = 3.55
MM1 pKa = 7.42SRR3 pKa = 11.84IRR5 pKa = 11.84NMMVTIYY12 pKa = 10.66FEE14 pKa = 6.12DD15 pKa = 3.93NGCPDD20 pKa = 3.81PPEE23 pKa = 4.35IRR25 pKa = 11.84AMLGALPNLRR35 pKa = 11.84YY36 pKa = 10.37YY37 pKa = 9.77IGQMEE42 pKa = 4.61KK43 pKa = 10.58CPSTGRR49 pKa = 11.84FHH51 pKa = 6.11FQGYY55 pKa = 9.81IEE57 pKa = 5.3FDD59 pKa = 2.9SAMYY63 pKa = 9.63VKK65 pKa = 10.47KK66 pKa = 10.73ISAQLNNCHH75 pKa = 6.17IEE77 pKa = 4.0ARR79 pKa = 11.84KK80 pKa = 8.34GTQLQAIAYY89 pKa = 6.89CKK91 pKa = 9.78KK92 pKa = 9.9HH93 pKa = 4.86EE94 pKa = 4.23TRR96 pKa = 11.84VGNWIEE102 pKa = 3.73YY103 pKa = 10.0GEE105 pKa = 4.24RR106 pKa = 11.84AKK108 pKa = 10.87QGTRR112 pKa = 11.84SDD114 pKa = 3.88LDD116 pKa = 3.94DD117 pKa = 3.8VAEE120 pKa = 4.82CIYY123 pKa = 10.42GGKK126 pKa = 9.72SIRR129 pKa = 11.84EE130 pKa = 3.94IQEE133 pKa = 3.67EE134 pKa = 4.33HH135 pKa = 6.23PRR137 pKa = 11.84AYY139 pKa = 10.25MMYY142 pKa = 10.26SKK144 pKa = 10.62GIKK147 pKa = 9.62DD148 pKa = 4.06LYY150 pKa = 8.66YY151 pKa = 9.6TVNNIEE157 pKa = 4.45PRR159 pKa = 11.84SEE161 pKa = 4.11PPVVIYY167 pKa = 10.35CYY169 pKa = 10.62GQTGTGKK176 pKa = 9.6SRR178 pKa = 11.84YY179 pKa = 8.9AKK181 pKa = 9.73RR182 pKa = 11.84YY183 pKa = 8.23SDD185 pKa = 3.43NVYY188 pKa = 10.13FKK190 pKa = 11.5NMSCKK195 pKa = 9.16WWDD198 pKa = 3.72GYY200 pKa = 11.35NGQDD204 pKa = 3.02VVVFDD209 pKa = 5.97DD210 pKa = 4.16IRR212 pKa = 11.84GDD214 pKa = 3.4TFKK217 pKa = 11.07YY218 pKa = 10.44HH219 pKa = 6.1EE220 pKa = 5.06LLRR223 pKa = 11.84ILDD226 pKa = 3.37RR227 pKa = 11.84YY228 pKa = 9.12VRR230 pKa = 11.84HH231 pKa = 6.11EE232 pKa = 4.47CDD234 pKa = 3.26SPNAPMVGIPCMLNIRR250 pKa = 11.84GAASTSIRR258 pKa = 11.84LL259 pKa = 3.55
Molecular weight: 29.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMJ6|A0A0C5IMJ6_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 20 OX=1618244 PE=4 SV=1
MM1 pKa = 7.57AFIPEE6 pKa = 5.17IIPEE10 pKa = 3.95IPEE13 pKa = 4.22IIEE16 pKa = 4.24GGEE19 pKa = 4.26ATWSAGRR26 pKa = 11.84AAVEE30 pKa = 3.63AWRR33 pKa = 11.84AGRR36 pKa = 11.84NLQAARR42 pKa = 11.84HH43 pKa = 5.24ALEE46 pKa = 3.92AARR49 pKa = 11.84RR50 pKa = 11.84AGTGIQAAEE59 pKa = 3.78RR60 pKa = 11.84TYY62 pKa = 10.85RR63 pKa = 11.84AAQRR67 pKa = 11.84HH68 pKa = 5.16YY69 pKa = 11.01NATHH73 pKa = 6.57HH74 pKa = 6.51NFGPTPPPNSPSGAPPISPASGPRR98 pKa = 11.84PTRR101 pKa = 11.84LRR103 pKa = 11.84IPRR106 pKa = 11.84TARR109 pKa = 11.84KK110 pKa = 10.13KK111 pKa = 9.71SMQVVTYY118 pKa = 9.35KK119 pKa = 10.63YY120 pKa = 9.95SRR122 pKa = 11.84GKK124 pKa = 8.96MRR126 pKa = 11.84YY127 pKa = 8.24KK128 pKa = 10.19KK129 pKa = 10.27SYY131 pKa = 8.33RR132 pKa = 11.84KK133 pKa = 9.82KK134 pKa = 10.13RR135 pKa = 11.84GRR137 pKa = 11.84KK138 pKa = 8.35YY139 pKa = 10.35KK140 pKa = 10.11RR141 pKa = 11.84KK142 pKa = 8.18TSATVPRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84NRR153 pKa = 11.84LRR155 pKa = 11.84GYY157 pKa = 10.45HH158 pKa = 6.07KK159 pKa = 10.94GKK161 pKa = 9.6GQMMGPTRR169 pKa = 11.84AAPRR173 pKa = 11.84YY174 pKa = 8.86QLVSNGWDD182 pKa = 3.2FDD184 pKa = 4.1YY185 pKa = 10.46PYY187 pKa = 9.74MNSGDD192 pKa = 3.98KK193 pKa = 9.66VTLEE197 pKa = 3.83LTQNNGALQLVKK209 pKa = 10.68YY210 pKa = 9.93IYY212 pKa = 10.97GKK214 pKa = 10.37FAFGWGSMGMEE225 pKa = 4.24VGSEE229 pKa = 4.01IGTHH233 pKa = 5.94AYY235 pKa = 9.04LWKK238 pKa = 10.42GVKK241 pKa = 9.73FVINYY246 pKa = 7.48NIPVGLGDD254 pKa = 3.78FKK256 pKa = 11.32IEE258 pKa = 4.04FFLCARR264 pKa = 11.84AVDD267 pKa = 4.0VDD269 pKa = 4.73SNPSLVLGEE278 pKa = 4.28SPGVKK283 pKa = 9.34VVPNRR288 pKa = 11.84VMLKK292 pKa = 8.22KK293 pKa = 10.28QSYY296 pKa = 9.1KK297 pKa = 10.62VFMHH301 pKa = 6.72RR302 pKa = 11.84KK303 pKa = 7.48YY304 pKa = 11.47NMRR307 pKa = 11.84FTASRR312 pKa = 11.84DD313 pKa = 3.74TIIGSDD319 pKa = 3.82PEE321 pKa = 4.32VTKK324 pKa = 10.7AAYY327 pKa = 9.34IKK329 pKa = 10.62GGNFSPHH336 pKa = 6.86CMFKK340 pKa = 11.0NPVTVRR346 pKa = 11.84FVKK349 pKa = 9.26HH350 pKa = 5.37TGIEE354 pKa = 4.05YY355 pKa = 11.01LMLGNNSTRR364 pKa = 11.84RR365 pKa = 11.84FPIDD369 pKa = 3.1SLMMVVRR376 pKa = 11.84ILPVLRR382 pKa = 11.84GSTTSGSEE390 pKa = 4.0AVFKK394 pKa = 10.52LASFAYY400 pKa = 7.88VTKK403 pKa = 9.72EE404 pKa = 3.91VPVLYY409 pKa = 10.73VV410 pKa = 3.02
MM1 pKa = 7.57AFIPEE6 pKa = 5.17IIPEE10 pKa = 3.95IPEE13 pKa = 4.22IIEE16 pKa = 4.24GGEE19 pKa = 4.26ATWSAGRR26 pKa = 11.84AAVEE30 pKa = 3.63AWRR33 pKa = 11.84AGRR36 pKa = 11.84NLQAARR42 pKa = 11.84HH43 pKa = 5.24ALEE46 pKa = 3.92AARR49 pKa = 11.84RR50 pKa = 11.84AGTGIQAAEE59 pKa = 3.78RR60 pKa = 11.84TYY62 pKa = 10.85RR63 pKa = 11.84AAQRR67 pKa = 11.84HH68 pKa = 5.16YY69 pKa = 11.01NATHH73 pKa = 6.57HH74 pKa = 6.51NFGPTPPPNSPSGAPPISPASGPRR98 pKa = 11.84PTRR101 pKa = 11.84LRR103 pKa = 11.84IPRR106 pKa = 11.84TARR109 pKa = 11.84KK110 pKa = 10.13KK111 pKa = 9.71SMQVVTYY118 pKa = 9.35KK119 pKa = 10.63YY120 pKa = 9.95SRR122 pKa = 11.84GKK124 pKa = 8.96MRR126 pKa = 11.84YY127 pKa = 8.24KK128 pKa = 10.19KK129 pKa = 10.27SYY131 pKa = 8.33RR132 pKa = 11.84KK133 pKa = 9.82KK134 pKa = 10.13RR135 pKa = 11.84GRR137 pKa = 11.84KK138 pKa = 8.35YY139 pKa = 10.35KK140 pKa = 10.11RR141 pKa = 11.84KK142 pKa = 8.18TSATVPRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84NRR153 pKa = 11.84LRR155 pKa = 11.84GYY157 pKa = 10.45HH158 pKa = 6.07KK159 pKa = 10.94GKK161 pKa = 9.6GQMMGPTRR169 pKa = 11.84AAPRR173 pKa = 11.84YY174 pKa = 8.86QLVSNGWDD182 pKa = 3.2FDD184 pKa = 4.1YY185 pKa = 10.46PYY187 pKa = 9.74MNSGDD192 pKa = 3.98KK193 pKa = 9.66VTLEE197 pKa = 3.83LTQNNGALQLVKK209 pKa = 10.68YY210 pKa = 9.93IYY212 pKa = 10.97GKK214 pKa = 10.37FAFGWGSMGMEE225 pKa = 4.24VGSEE229 pKa = 4.01IGTHH233 pKa = 5.94AYY235 pKa = 9.04LWKK238 pKa = 10.42GVKK241 pKa = 9.73FVINYY246 pKa = 7.48NIPVGLGDD254 pKa = 3.78FKK256 pKa = 11.32IEE258 pKa = 4.04FFLCARR264 pKa = 11.84AVDD267 pKa = 4.0VDD269 pKa = 4.73SNPSLVLGEE278 pKa = 4.28SPGVKK283 pKa = 9.34VVPNRR288 pKa = 11.84VMLKK292 pKa = 8.22KK293 pKa = 10.28QSYY296 pKa = 9.1KK297 pKa = 10.62VFMHH301 pKa = 6.72RR302 pKa = 11.84KK303 pKa = 7.48YY304 pKa = 11.47NMRR307 pKa = 11.84FTASRR312 pKa = 11.84DD313 pKa = 3.74TIIGSDD319 pKa = 3.82PEE321 pKa = 4.32VTKK324 pKa = 10.7AAYY327 pKa = 9.34IKK329 pKa = 10.62GGNFSPHH336 pKa = 6.86CMFKK340 pKa = 11.0NPVTVRR346 pKa = 11.84FVKK349 pKa = 9.26HH350 pKa = 5.37TGIEE354 pKa = 4.05YY355 pKa = 11.01LMLGNNSTRR364 pKa = 11.84RR365 pKa = 11.84FPIDD369 pKa = 3.1SLMMVVRR376 pKa = 11.84ILPVLRR382 pKa = 11.84GSTTSGSEE390 pKa = 4.0AVFKK394 pKa = 10.52LASFAYY400 pKa = 7.88VTKK403 pKa = 9.72EE404 pKa = 3.91VPVLYY409 pKa = 10.73VV410 pKa = 3.02
Molecular weight: 46.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
669 |
259 |
410 |
334.5 |
38.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.324 ± 1.108 | 1.644 ± 1.057 |
3.587 ± 1.272 | 4.933 ± 0.719 |
3.587 ± 0.511 | 8.371 ± 0.374 |
2.242 ± 0.043 | 5.979 ± 1.006 |
6.726 ± 0.54 | 5.082 ± 0.259 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.886 ± 0.208 | 4.634 ± 0.223 |
5.979 ± 0.777 | 2.691 ± 0.453 |
8.52 ± 0.461 | 5.979 ± 0.331 |
4.933 ± 0.619 | 6.577 ± 0.899 |
1.196 ± 0.022 | 6.129 ± 0.92 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
