
Lodeiro virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins
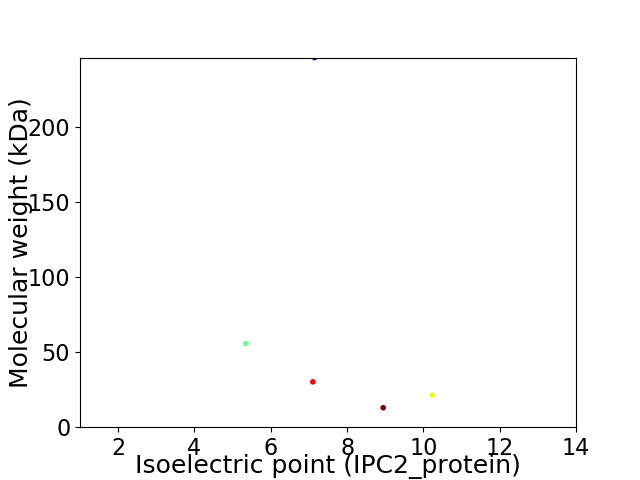
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D9CFP5|A0A1D9CFP5_9VIRU ORF3 OS=Lodeiro virus OX=1911102 PE=4 SV=1
MM1 pKa = 7.0MVSLTGCFLLIRR13 pKa = 11.84LLIILKK19 pKa = 9.99IPVVLEE25 pKa = 3.82SKK27 pKa = 9.92YY28 pKa = 11.25VKK30 pKa = 10.24DD31 pKa = 3.41LHH33 pKa = 6.84PYY35 pKa = 8.63IANFSQIHH43 pKa = 5.63SLLVGRR49 pKa = 11.84RR50 pKa = 11.84AYY52 pKa = 10.94LEE54 pKa = 4.56DD55 pKa = 4.32FNFLQRR61 pKa = 11.84NIIMDD66 pKa = 4.43VKK68 pKa = 8.12PHH70 pKa = 6.72RR71 pKa = 11.84YY72 pKa = 9.35VMQDD76 pKa = 2.8RR77 pKa = 11.84DD78 pKa = 3.13EE79 pKa = 4.25WNIDD83 pKa = 3.06NCIKK87 pKa = 10.66FIEE90 pKa = 4.36THH92 pKa = 5.96SLIPDD97 pKa = 4.08DD98 pKa = 4.45CQSILGDD105 pKa = 3.65EE106 pKa = 4.64CEE108 pKa = 4.17GATFKK113 pKa = 11.11SIEE116 pKa = 4.48TKK118 pKa = 8.79WHH120 pKa = 5.66GLQLGICHH128 pKa = 6.51PTPNCTFQDD137 pKa = 3.68FDD139 pKa = 4.31QSYY142 pKa = 8.76QCGNAKK148 pKa = 9.61YY149 pKa = 10.28VLYY152 pKa = 10.68HH153 pKa = 7.25EE154 pKa = 4.54YY155 pKa = 10.38TFDD158 pKa = 2.94IPYY161 pKa = 10.43VVFVPVKK168 pKa = 10.38RR169 pKa = 11.84NNKK172 pKa = 5.79VTWTAILEE180 pKa = 4.24YY181 pKa = 10.96NEE183 pKa = 4.5EE184 pKa = 4.43DD185 pKa = 3.21LSKK188 pKa = 10.21YY189 pKa = 9.81QSPGSFSLKK198 pKa = 10.29SHH200 pKa = 6.14MQQSSYY206 pKa = 10.93YY207 pKa = 9.39IKK209 pKa = 10.63QLFDD213 pKa = 3.58RR214 pKa = 11.84SLPDD218 pKa = 3.27DD219 pKa = 3.91VKK221 pKa = 11.49YY222 pKa = 8.2EE223 pKa = 4.06TQIQQTEE230 pKa = 4.42CVEE233 pKa = 4.16SLPFYY238 pKa = 10.86DD239 pKa = 5.07ADD241 pKa = 3.69GFYY244 pKa = 10.69HH245 pKa = 7.24EE246 pKa = 5.46NIAYY250 pKa = 10.35LNHH253 pKa = 6.5DD254 pKa = 4.28WKK256 pKa = 11.16RR257 pKa = 11.84MCFSLAPQGSYY268 pKa = 10.73CDD270 pKa = 3.87SLWWVDD276 pKa = 4.17DD277 pKa = 4.1YY278 pKa = 11.83FDD280 pKa = 3.87KK281 pKa = 9.15TTLVKK286 pKa = 10.66ALLEE290 pKa = 4.47EE291 pKa = 4.3IPWYY295 pKa = 9.82IVYY298 pKa = 10.4HH299 pKa = 6.63DD300 pKa = 4.36WNSLKK305 pKa = 10.47QSIKK309 pKa = 10.86DD310 pKa = 3.53QVKK313 pKa = 8.83TDD315 pKa = 3.85CEE317 pKa = 4.18IEE319 pKa = 3.99NDD321 pKa = 2.72IHH323 pKa = 7.15LRR325 pKa = 11.84WNVTHH330 pKa = 7.5CYY332 pKa = 10.05QGDD335 pKa = 4.56FKK337 pKa = 11.42CMYY340 pKa = 10.65GPFPCSDD347 pKa = 3.11SANRR351 pKa = 11.84VSKK354 pKa = 10.87CLDD357 pKa = 3.31VEE359 pKa = 4.39EE360 pKa = 5.55HH361 pKa = 5.54FTSPVTFHH369 pKa = 6.34AVKK372 pKa = 10.41VVRR375 pKa = 11.84DD376 pKa = 3.79VVKK379 pKa = 10.93AILPYY384 pKa = 10.23IKK386 pKa = 9.85TIFTTVLGIIVDD398 pKa = 3.8VFNSVLVQLGLVFMLLTTASILVMVIKK425 pKa = 10.74FGLRR429 pKa = 11.84YY430 pKa = 9.54LDD432 pKa = 3.51SSLILLTLTLLLSTLGIDD450 pKa = 4.36LKK452 pKa = 11.12NSYY455 pKa = 10.24FISCILVLTASKK467 pKa = 10.36YY468 pKa = 9.62SKK470 pKa = 10.48VLEE473 pKa = 4.38RR474 pKa = 11.84YY475 pKa = 9.41VKK477 pKa = 10.58EE478 pKa = 4.02NN479 pKa = 3.03
MM1 pKa = 7.0MVSLTGCFLLIRR13 pKa = 11.84LLIILKK19 pKa = 9.99IPVVLEE25 pKa = 3.82SKK27 pKa = 9.92YY28 pKa = 11.25VKK30 pKa = 10.24DD31 pKa = 3.41LHH33 pKa = 6.84PYY35 pKa = 8.63IANFSQIHH43 pKa = 5.63SLLVGRR49 pKa = 11.84RR50 pKa = 11.84AYY52 pKa = 10.94LEE54 pKa = 4.56DD55 pKa = 4.32FNFLQRR61 pKa = 11.84NIIMDD66 pKa = 4.43VKK68 pKa = 8.12PHH70 pKa = 6.72RR71 pKa = 11.84YY72 pKa = 9.35VMQDD76 pKa = 2.8RR77 pKa = 11.84DD78 pKa = 3.13EE79 pKa = 4.25WNIDD83 pKa = 3.06NCIKK87 pKa = 10.66FIEE90 pKa = 4.36THH92 pKa = 5.96SLIPDD97 pKa = 4.08DD98 pKa = 4.45CQSILGDD105 pKa = 3.65EE106 pKa = 4.64CEE108 pKa = 4.17GATFKK113 pKa = 11.11SIEE116 pKa = 4.48TKK118 pKa = 8.79WHH120 pKa = 5.66GLQLGICHH128 pKa = 6.51PTPNCTFQDD137 pKa = 3.68FDD139 pKa = 4.31QSYY142 pKa = 8.76QCGNAKK148 pKa = 9.61YY149 pKa = 10.28VLYY152 pKa = 10.68HH153 pKa = 7.25EE154 pKa = 4.54YY155 pKa = 10.38TFDD158 pKa = 2.94IPYY161 pKa = 10.43VVFVPVKK168 pKa = 10.38RR169 pKa = 11.84NNKK172 pKa = 5.79VTWTAILEE180 pKa = 4.24YY181 pKa = 10.96NEE183 pKa = 4.5EE184 pKa = 4.43DD185 pKa = 3.21LSKK188 pKa = 10.21YY189 pKa = 9.81QSPGSFSLKK198 pKa = 10.29SHH200 pKa = 6.14MQQSSYY206 pKa = 10.93YY207 pKa = 9.39IKK209 pKa = 10.63QLFDD213 pKa = 3.58RR214 pKa = 11.84SLPDD218 pKa = 3.27DD219 pKa = 3.91VKK221 pKa = 11.49YY222 pKa = 8.2EE223 pKa = 4.06TQIQQTEE230 pKa = 4.42CVEE233 pKa = 4.16SLPFYY238 pKa = 10.86DD239 pKa = 5.07ADD241 pKa = 3.69GFYY244 pKa = 10.69HH245 pKa = 7.24EE246 pKa = 5.46NIAYY250 pKa = 10.35LNHH253 pKa = 6.5DD254 pKa = 4.28WKK256 pKa = 11.16RR257 pKa = 11.84MCFSLAPQGSYY268 pKa = 10.73CDD270 pKa = 3.87SLWWVDD276 pKa = 4.17DD277 pKa = 4.1YY278 pKa = 11.83FDD280 pKa = 3.87KK281 pKa = 9.15TTLVKK286 pKa = 10.66ALLEE290 pKa = 4.47EE291 pKa = 4.3IPWYY295 pKa = 9.82IVYY298 pKa = 10.4HH299 pKa = 6.63DD300 pKa = 4.36WNSLKK305 pKa = 10.47QSIKK309 pKa = 10.86DD310 pKa = 3.53QVKK313 pKa = 8.83TDD315 pKa = 3.85CEE317 pKa = 4.18IEE319 pKa = 3.99NDD321 pKa = 2.72IHH323 pKa = 7.15LRR325 pKa = 11.84WNVTHH330 pKa = 7.5CYY332 pKa = 10.05QGDD335 pKa = 4.56FKK337 pKa = 11.42CMYY340 pKa = 10.65GPFPCSDD347 pKa = 3.11SANRR351 pKa = 11.84VSKK354 pKa = 10.87CLDD357 pKa = 3.31VEE359 pKa = 4.39EE360 pKa = 5.55HH361 pKa = 5.54FTSPVTFHH369 pKa = 6.34AVKK372 pKa = 10.41VVRR375 pKa = 11.84DD376 pKa = 3.79VVKK379 pKa = 10.93AILPYY384 pKa = 10.23IKK386 pKa = 9.85TIFTTVLGIIVDD398 pKa = 3.8VFNSVLVQLGLVFMLLTTASILVMVIKK425 pKa = 10.74FGLRR429 pKa = 11.84YY430 pKa = 9.54LDD432 pKa = 3.51SSLILLTLTLLLSTLGIDD450 pKa = 4.36LKK452 pKa = 11.12NSYY455 pKa = 10.24FISCILVLTASKK467 pKa = 10.36YY468 pKa = 9.62SKK470 pKa = 10.48VLEE473 pKa = 4.38RR474 pKa = 11.84YY475 pKa = 9.41VKK477 pKa = 10.58EE478 pKa = 4.02NN479 pKa = 3.03
Molecular weight: 55.76 kDa
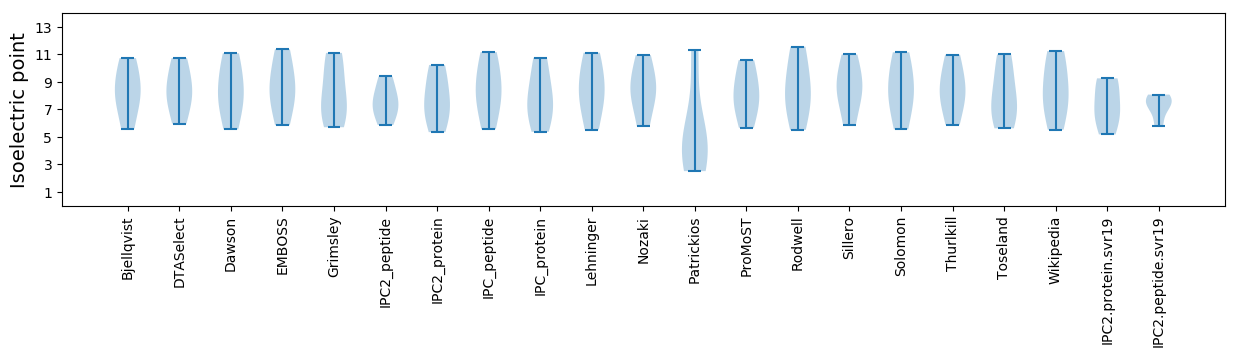
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D9CFR9|A0A1D9CFR9_9VIRU ORF5 OS=Lodeiro virus OX=1911102 PE=4 SV=1
MM1 pKa = 7.38AKK3 pKa = 10.08KK4 pKa = 9.97YY5 pKa = 6.32QTRR8 pKa = 11.84RR9 pKa = 11.84TRR11 pKa = 11.84LTAVGTARR19 pKa = 11.84KK20 pKa = 9.58AIRR23 pKa = 11.84KK24 pKa = 6.76TKK26 pKa = 9.0TKK28 pKa = 10.52SPKK31 pKa = 8.07TQVLVVSNGKK41 pKa = 9.26RR42 pKa = 11.84KK43 pKa = 9.3SRR45 pKa = 11.84KK46 pKa = 9.08RR47 pKa = 11.84EE48 pKa = 3.69NKK50 pKa = 9.72KK51 pKa = 10.49LVGVEE56 pKa = 4.07GAYY59 pKa = 10.43HH60 pKa = 6.04SVVKK64 pKa = 10.39VLNEE68 pKa = 4.1PYY70 pKa = 10.02TLILIGLALLVIVNYY85 pKa = 10.31QNSKK89 pKa = 8.12TDD91 pKa = 3.48NVVTKK96 pKa = 10.73LADD99 pKa = 3.78TIGNQTSLGIWMKK112 pKa = 11.37ANVAKK117 pKa = 10.93VIGLAIMAPSVFTSPKK133 pKa = 9.75SIRR136 pKa = 11.84VPLAIGSLALVYY148 pKa = 10.03LVKK151 pKa = 10.67ALSLWNYY158 pKa = 9.02FSIAIAIRR166 pKa = 11.84VFFKK170 pKa = 11.29VKK172 pKa = 10.05DD173 pKa = 3.51QNIRR177 pKa = 11.84LIVFGLAFAMYY188 pKa = 10.13YY189 pKa = 9.96IGASS193 pKa = 3.3
MM1 pKa = 7.38AKK3 pKa = 10.08KK4 pKa = 9.97YY5 pKa = 6.32QTRR8 pKa = 11.84RR9 pKa = 11.84TRR11 pKa = 11.84LTAVGTARR19 pKa = 11.84KK20 pKa = 9.58AIRR23 pKa = 11.84KK24 pKa = 6.76TKK26 pKa = 9.0TKK28 pKa = 10.52SPKK31 pKa = 8.07TQVLVVSNGKK41 pKa = 9.26RR42 pKa = 11.84KK43 pKa = 9.3SRR45 pKa = 11.84KK46 pKa = 9.08RR47 pKa = 11.84EE48 pKa = 3.69NKK50 pKa = 9.72KK51 pKa = 10.49LVGVEE56 pKa = 4.07GAYY59 pKa = 10.43HH60 pKa = 6.04SVVKK64 pKa = 10.39VLNEE68 pKa = 4.1PYY70 pKa = 10.02TLILIGLALLVIVNYY85 pKa = 10.31QNSKK89 pKa = 8.12TDD91 pKa = 3.48NVVTKK96 pKa = 10.73LADD99 pKa = 3.78TIGNQTSLGIWMKK112 pKa = 11.37ANVAKK117 pKa = 10.93VIGLAIMAPSVFTSPKK133 pKa = 9.75SIRR136 pKa = 11.84VPLAIGSLALVYY148 pKa = 10.03LVKK151 pKa = 10.67ALSLWNYY158 pKa = 9.02FSIAIAIRR166 pKa = 11.84VFFKK170 pKa = 11.29VKK172 pKa = 10.05DD173 pKa = 3.51QNIRR177 pKa = 11.84LIVFGLAFAMYY188 pKa = 10.13YY189 pKa = 9.96IGASS193 pKa = 3.3
Molecular weight: 21.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3157 |
109 |
2116 |
631.4 |
73.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.561 ± 0.773 | 1.742 ± 0.414 |
5.86 ± 0.824 | 7.507 ± 1.14 |
4.435 ± 0.537 | 4.53 ± 0.388 |
2.787 ± 0.319 | 6.525 ± 0.388 |
8.679 ± 0.877 | 8.933 ± 0.704 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 0.325 | 4.023 ± 0.401 |
3.389 ± 0.15 | 3.833 ± 0.327 |
5.512 ± 0.673 | 6.937 ± 0.437 |
4.498 ± 0.639 | 7.697 ± 0.497 |
1.235 ± 0.152 | 4.72 ± 0.404 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
