
Cohaesibacter celericrescens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Cohaesibacteraceae; Cohaesibacter
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
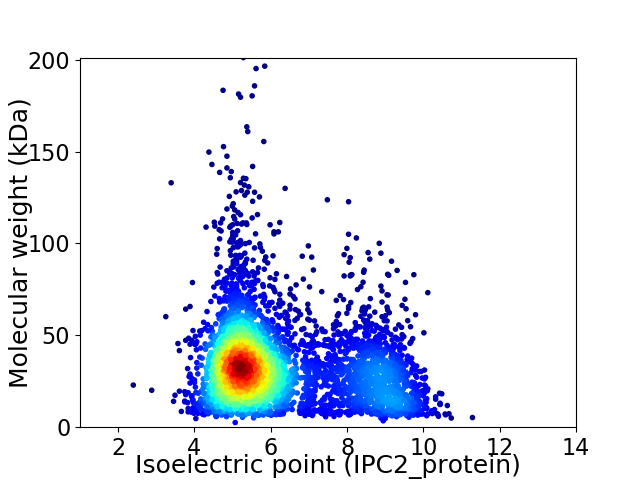
Virtual 2D-PAGE plot for 4355 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N5XKE9|A0A2N5XKE9_9RHIZ Short-chain dehydrogenase OS=Cohaesibacter celericrescens OX=2067669 GN=C0081_22070 PE=3 SV=1
MM1 pKa = 7.44NIKK4 pKa = 10.47SIILGSAAALVAGGAAQAADD24 pKa = 4.15LPVAEE29 pKa = 4.66PVDD32 pKa = 3.96YY33 pKa = 11.46VKK35 pKa = 11.15VCDD38 pKa = 4.56AYY40 pKa = 9.79GAGYY44 pKa = 9.68FFIPGTDD51 pKa = 3.07TCLKK55 pKa = 10.35IGGKK59 pKa = 8.6VLFGAEE65 pKa = 3.98SAGFNSDD72 pKa = 3.01VARR75 pKa = 11.84SDD77 pKa = 3.45NLVNFYY83 pKa = 10.8TEE85 pKa = 3.99TTFKK89 pKa = 9.9FTARR93 pKa = 11.84EE94 pKa = 3.91EE95 pKa = 4.65TEE97 pKa = 4.73LGTLTAHH104 pKa = 6.61MEE106 pKa = 4.04FDD108 pKa = 3.96DD109 pKa = 4.83TGEE112 pKa = 4.7DD113 pKa = 3.76DD114 pKa = 4.74LGEE117 pKa = 4.18GTAIDD122 pKa = 3.68KK123 pKa = 10.98AYY125 pKa = 8.35ITLGGFYY132 pKa = 10.48AGLTDD137 pKa = 4.82SLLSFNAGYY146 pKa = 10.5GYY148 pKa = 10.71DD149 pKa = 3.92DD150 pKa = 4.36YY151 pKa = 12.12GVLDD155 pKa = 3.72VDD157 pKa = 5.63LNTIGYY163 pKa = 8.17KK164 pKa = 10.37AEE166 pKa = 4.0MGNGVTAAIALEE178 pKa = 4.54EE179 pKa = 4.12YY180 pKa = 9.74TGNEE184 pKa = 3.85TGAGTSMPAVVAKK197 pKa = 10.27LSIAQGWGSAEE208 pKa = 3.84AGGSVFQVRR217 pKa = 11.84YY218 pKa = 9.77QDD220 pKa = 3.31AAYY223 pKa = 8.5DD224 pKa = 3.68TDD226 pKa = 3.18YY227 pKa = 11.42GYY229 pKa = 11.61AFGGQAKK236 pKa = 9.81FNVVEE241 pKa = 4.1DD242 pKa = 4.15LKK244 pKa = 11.36FGVAGGYY251 pKa = 6.5TAGGSWLEE259 pKa = 4.06EE260 pKa = 4.0SAVTGKK266 pKa = 10.94LNTGWAASAGLEE278 pKa = 4.15YY279 pKa = 11.08AFADD283 pKa = 3.32NFNFYY288 pKa = 10.96VDD290 pKa = 3.61AGYY293 pKa = 10.71KK294 pKa = 10.22AFNDD298 pKa = 4.37RR299 pKa = 11.84SGTDD303 pKa = 3.14DD304 pKa = 3.39WEE306 pKa = 4.52GWGVSAMAEE315 pKa = 4.01YY316 pKa = 10.5FPVKK320 pKa = 10.12NLSIAATVGYY330 pKa = 9.95QKK332 pKa = 10.35IDD334 pKa = 3.39FDD336 pKa = 4.49TEE338 pKa = 3.88TTANQDD344 pKa = 3.11WDD346 pKa = 3.97DD347 pKa = 3.76VAAKK351 pKa = 10.65LEE353 pKa = 4.03IEE355 pKa = 4.26RR356 pKa = 11.84TFF358 pKa = 3.33
MM1 pKa = 7.44NIKK4 pKa = 10.47SIILGSAAALVAGGAAQAADD24 pKa = 4.15LPVAEE29 pKa = 4.66PVDD32 pKa = 3.96YY33 pKa = 11.46VKK35 pKa = 11.15VCDD38 pKa = 4.56AYY40 pKa = 9.79GAGYY44 pKa = 9.68FFIPGTDD51 pKa = 3.07TCLKK55 pKa = 10.35IGGKK59 pKa = 8.6VLFGAEE65 pKa = 3.98SAGFNSDD72 pKa = 3.01VARR75 pKa = 11.84SDD77 pKa = 3.45NLVNFYY83 pKa = 10.8TEE85 pKa = 3.99TTFKK89 pKa = 9.9FTARR93 pKa = 11.84EE94 pKa = 3.91EE95 pKa = 4.65TEE97 pKa = 4.73LGTLTAHH104 pKa = 6.61MEE106 pKa = 4.04FDD108 pKa = 3.96DD109 pKa = 4.83TGEE112 pKa = 4.7DD113 pKa = 3.76DD114 pKa = 4.74LGEE117 pKa = 4.18GTAIDD122 pKa = 3.68KK123 pKa = 10.98AYY125 pKa = 8.35ITLGGFYY132 pKa = 10.48AGLTDD137 pKa = 4.82SLLSFNAGYY146 pKa = 10.5GYY148 pKa = 10.71DD149 pKa = 3.92DD150 pKa = 4.36YY151 pKa = 12.12GVLDD155 pKa = 3.72VDD157 pKa = 5.63LNTIGYY163 pKa = 8.17KK164 pKa = 10.37AEE166 pKa = 4.0MGNGVTAAIALEE178 pKa = 4.54EE179 pKa = 4.12YY180 pKa = 9.74TGNEE184 pKa = 3.85TGAGTSMPAVVAKK197 pKa = 10.27LSIAQGWGSAEE208 pKa = 3.84AGGSVFQVRR217 pKa = 11.84YY218 pKa = 9.77QDD220 pKa = 3.31AAYY223 pKa = 8.5DD224 pKa = 3.68TDD226 pKa = 3.18YY227 pKa = 11.42GYY229 pKa = 11.61AFGGQAKK236 pKa = 9.81FNVVEE241 pKa = 4.1DD242 pKa = 4.15LKK244 pKa = 11.36FGVAGGYY251 pKa = 6.5TAGGSWLEE259 pKa = 4.06EE260 pKa = 4.0SAVTGKK266 pKa = 10.94LNTGWAASAGLEE278 pKa = 4.15YY279 pKa = 11.08AFADD283 pKa = 3.32NFNFYY288 pKa = 10.96VDD290 pKa = 3.61AGYY293 pKa = 10.71KK294 pKa = 10.22AFNDD298 pKa = 4.37RR299 pKa = 11.84SGTDD303 pKa = 3.14DD304 pKa = 3.39WEE306 pKa = 4.52GWGVSAMAEE315 pKa = 4.01YY316 pKa = 10.5FPVKK320 pKa = 10.12NLSIAATVGYY330 pKa = 9.95QKK332 pKa = 10.35IDD334 pKa = 3.39FDD336 pKa = 4.49TEE338 pKa = 3.88TTANQDD344 pKa = 3.11WDD346 pKa = 3.97DD347 pKa = 3.76VAAKK351 pKa = 10.65LEE353 pKa = 4.03IEE355 pKa = 4.26RR356 pKa = 11.84TFF358 pKa = 3.33
Molecular weight: 37.89 kDa
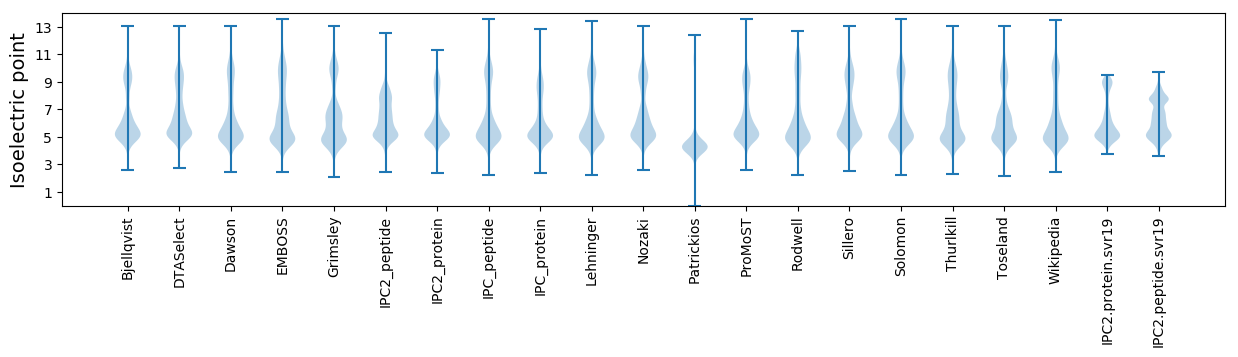
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N5XP47|A0A2N5XP47_9RHIZ DUF547 domain-containing protein OS=Cohaesibacter celericrescens OX=2067669 GN=C0081_14920 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.43NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.43NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1385776 |
24 |
1854 |
318.2 |
34.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.42 ± 0.04 | 0.956 ± 0.011 |
5.919 ± 0.034 | 5.696 ± 0.036 |
4.024 ± 0.026 | 7.885 ± 0.034 |
2.109 ± 0.019 | 6.162 ± 0.033 |
4.492 ± 0.029 | 10.172 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.834 ± 0.019 | 3.319 ± 0.02 |
4.463 ± 0.025 | 3.633 ± 0.023 |
5.476 ± 0.034 | 6.407 ± 0.029 |
5.337 ± 0.025 | 7.05 ± 0.029 |
1.244 ± 0.013 | 2.403 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
