
Agromyces binzhouensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
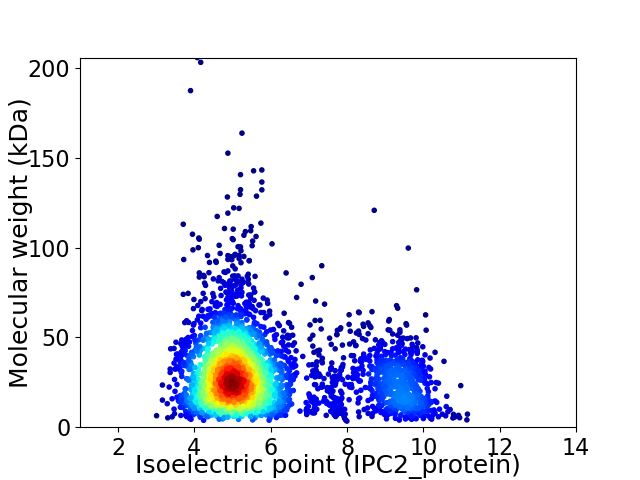
Virtual 2D-PAGE plot for 3584 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
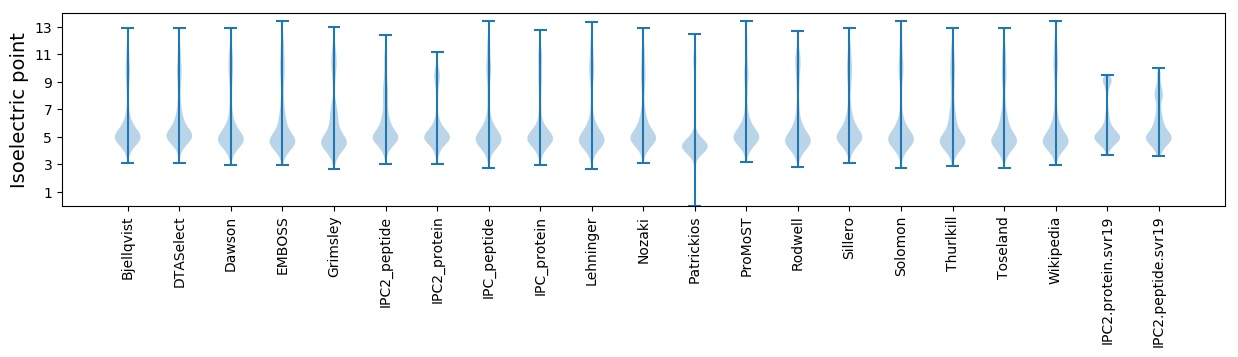
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2JWA0|A0A4Q2JWA0_9MICO ABC transporter permease OS=Agromyces binzhouensis OX=1817495 GN=ESO86_01300 PE=4 SV=1
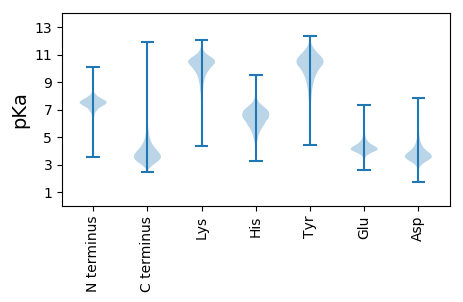
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84MIEE6 pKa = 3.88RR7 pKa = 11.84LRR9 pKa = 11.84GGEE12 pKa = 3.9RR13 pKa = 11.84GASAVLVGLMIVPLIGAAAIAVDD36 pKa = 3.98VGALYY41 pKa = 10.78AEE43 pKa = 5.14RR44 pKa = 11.84AAQQNGADD52 pKa = 3.86GAAMAVATACAADD65 pKa = 4.25EE66 pKa = 4.5SACGASAGSIASDD79 pKa = 3.6FVLQNSFISSPTALAPQIDD98 pKa = 4.21YY99 pKa = 10.54DD100 pKa = 4.2ANRR103 pKa = 11.84VTVTTDD109 pKa = 2.78NSVSHH114 pKa = 6.94PLASVLTGINSTVVQAVSSAEE135 pKa = 3.8WGSPNSGIVLPLALSLCEE153 pKa = 4.13FQDD156 pKa = 3.3PTLEE160 pKa = 3.71IRR162 pKa = 11.84VLIEE166 pKa = 3.31YY167 pKa = 9.34HH168 pKa = 4.74QTSEE172 pKa = 4.04NDD174 pKa = 3.48CTRR177 pKa = 11.84SDD179 pKa = 3.69GQPVEE184 pKa = 5.72GGFGWLDD191 pKa = 3.58LEE193 pKa = 5.05GGQCEE198 pKa = 4.27TFIDD202 pKa = 4.64LDD204 pKa = 3.83TGFVGSDD211 pKa = 3.3PGIDD215 pKa = 3.45PAAEE219 pKa = 3.76CAYY222 pKa = 10.65LFNDD226 pKa = 4.41LEE228 pKa = 4.91GEE230 pKa = 4.51TVLVPVYY237 pKa = 10.88DD238 pKa = 4.03GGNDD242 pKa = 3.09INGQNGEE249 pKa = 4.03FHH251 pKa = 7.23IMAFAAFVVTGWKK264 pKa = 8.41LTGGGPGVFNNPDD277 pKa = 4.16PLSPACVGACRR288 pKa = 11.84GIQGYY293 pKa = 6.83FTEE296 pKa = 4.68WVDD299 pKa = 5.68LGADD303 pKa = 3.73WEE305 pKa = 4.66VGGPDD310 pKa = 3.35FGVDD314 pKa = 3.23VVRR317 pKa = 11.84LVITDD322 pKa = 3.51SQLADD327 pKa = 4.0LLDD330 pKa = 3.7
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84MIEE6 pKa = 3.88RR7 pKa = 11.84LRR9 pKa = 11.84GGEE12 pKa = 3.9RR13 pKa = 11.84GASAVLVGLMIVPLIGAAAIAVDD36 pKa = 3.98VGALYY41 pKa = 10.78AEE43 pKa = 5.14RR44 pKa = 11.84AAQQNGADD52 pKa = 3.86GAAMAVATACAADD65 pKa = 4.25EE66 pKa = 4.5SACGASAGSIASDD79 pKa = 3.6FVLQNSFISSPTALAPQIDD98 pKa = 4.21YY99 pKa = 10.54DD100 pKa = 4.2ANRR103 pKa = 11.84VTVTTDD109 pKa = 2.78NSVSHH114 pKa = 6.94PLASVLTGINSTVVQAVSSAEE135 pKa = 3.8WGSPNSGIVLPLALSLCEE153 pKa = 4.13FQDD156 pKa = 3.3PTLEE160 pKa = 3.71IRR162 pKa = 11.84VLIEE166 pKa = 3.31YY167 pKa = 9.34HH168 pKa = 4.74QTSEE172 pKa = 4.04NDD174 pKa = 3.48CTRR177 pKa = 11.84SDD179 pKa = 3.69GQPVEE184 pKa = 5.72GGFGWLDD191 pKa = 3.58LEE193 pKa = 5.05GGQCEE198 pKa = 4.27TFIDD202 pKa = 4.64LDD204 pKa = 3.83TGFVGSDD211 pKa = 3.3PGIDD215 pKa = 3.45PAAEE219 pKa = 3.76CAYY222 pKa = 10.65LFNDD226 pKa = 4.41LEE228 pKa = 4.91GEE230 pKa = 4.51TVLVPVYY237 pKa = 10.88DD238 pKa = 4.03GGNDD242 pKa = 3.09INGQNGEE249 pKa = 4.03FHH251 pKa = 7.23IMAFAAFVVTGWKK264 pKa = 8.41LTGGGPGVFNNPDD277 pKa = 4.16PLSPACVGACRR288 pKa = 11.84GIQGYY293 pKa = 6.83FTEE296 pKa = 4.68WVDD299 pKa = 5.68LGADD303 pKa = 3.73WEE305 pKa = 4.66VGGPDD310 pKa = 3.35FGVDD314 pKa = 3.23VVRR317 pKa = 11.84LVITDD322 pKa = 3.51SQLADD327 pKa = 4.0LLDD330 pKa = 3.7
Molecular weight: 34.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2J4D3|A0A4Q2J4D3_9MICO Uncharacterized protein (Fragment) OS=Agromyces binzhouensis OX=1817495 GN=ESO86_17555 PE=4 SV=1
MM1 pKa = 7.64PGGVAAQAVRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84APRR30 pKa = 11.84CRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84AGTPRR46 pKa = 11.84GRR48 pKa = 11.84CGRR51 pKa = 11.84PASRR55 pKa = 11.84ARR57 pKa = 11.84RR58 pKa = 11.84SPRR61 pKa = 3.05
MM1 pKa = 7.64PGGVAAQAVRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84APRR30 pKa = 11.84CRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84AGTPRR46 pKa = 11.84GRR48 pKa = 11.84CGRR51 pKa = 11.84PASRR55 pKa = 11.84ARR57 pKa = 11.84RR58 pKa = 11.84SPRR61 pKa = 3.05
Molecular weight: 7.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1048746 |
32 |
1954 |
292.6 |
31.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.855 ± 0.059 | 0.47 ± 0.01 |
6.528 ± 0.034 | 6.093 ± 0.042 |
3.136 ± 0.025 | 9.203 ± 0.037 |
2.034 ± 0.02 | 4.33 ± 0.025 |
1.682 ± 0.033 | 9.954 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.749 ± 0.016 | 1.8 ± 0.021 |
5.453 ± 0.033 | 2.411 ± 0.024 |
7.767 ± 0.05 | 5.247 ± 0.026 |
5.595 ± 0.028 | 9.216 ± 0.044 |
1.528 ± 0.019 | 1.949 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
