
Bovine group B rotavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus B
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
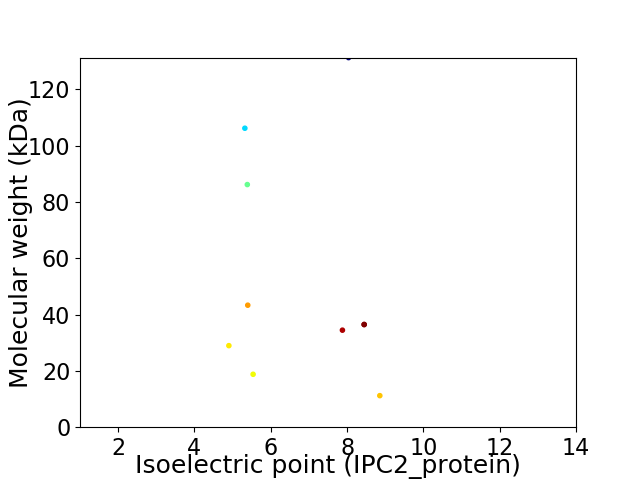
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
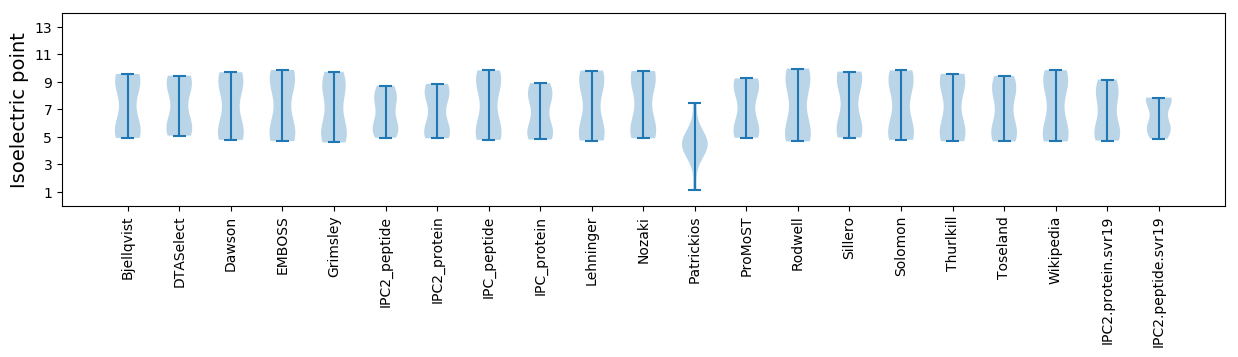
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6KF86|Q6KF86_9REOV Outer capsid glycoprotein VP7 OS=Bovine group B rotavirus OX=35334 GN=VP7 PE=3 SV=1
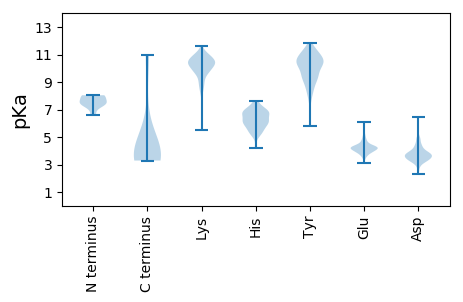
MM1 pKa = 8.02PLPLLLVLAACAKK14 pKa = 9.3AQLIITPISNPEE26 pKa = 3.61ICVLHH31 pKa = 6.92ANDD34 pKa = 4.38RR35 pKa = 11.84NVNTFGEE42 pKa = 4.27NFTNIFEE49 pKa = 4.87SYY51 pKa = 11.2NSVTLSFYY59 pKa = 11.1NYY61 pKa = 10.31DD62 pKa = 3.57SNNYY66 pKa = 9.78DD67 pKa = 3.8VIDD70 pKa = 3.82ILSKK74 pKa = 10.58KK75 pKa = 10.31DD76 pKa = 3.47YY77 pKa = 10.63SLCQILAIDD86 pKa = 4.34VIRR89 pKa = 11.84PEE91 pKa = 3.68MDD93 pKa = 4.33FITFLQSNNEE103 pKa = 3.75CSKK106 pKa = 10.69YY107 pKa = 10.16SRR109 pKa = 11.84QKK111 pKa = 9.79IHH113 pKa = 5.52YY114 pKa = 8.27QKK116 pKa = 10.11LTTGDD121 pKa = 2.86EE122 pKa = 3.97WFVYY126 pKa = 10.29SKK128 pKa = 11.0NLKK131 pKa = 9.78FCPLSDD137 pKa = 3.76NLIGLYY143 pKa = 10.43CDD145 pKa = 3.46TQVNGTYY152 pKa = 10.25FPLSNNEE159 pKa = 4.16RR160 pKa = 11.84YY161 pKa = 10.04DD162 pKa = 3.63VTDD165 pKa = 3.66LPEE168 pKa = 4.25FTEE171 pKa = 3.96MGYY174 pKa = 10.53VFYY177 pKa = 10.98SDD179 pKa = 4.25DD180 pKa = 4.04DD181 pKa = 4.4FYY183 pKa = 10.89ICKK186 pKa = 10.14RR187 pKa = 11.84VTEE190 pKa = 4.36EE191 pKa = 3.11KK192 pKa = 9.67WINYY196 pKa = 8.41HH197 pKa = 6.33LFYY200 pKa = 10.77RR201 pKa = 11.84EE202 pKa = 3.82YY203 pKa = 10.22SASGTVARR211 pKa = 11.84AISWDD216 pKa = 3.87NVWTGFKK223 pKa = 10.06TFAQVVYY230 pKa = 10.17KK231 pKa = 10.64ILDD234 pKa = 3.15IFFNNRR240 pKa = 11.84RR241 pKa = 11.84NFEE244 pKa = 3.84PRR246 pKa = 11.84AA247 pKa = 3.56
MM1 pKa = 8.02PLPLLLVLAACAKK14 pKa = 9.3AQLIITPISNPEE26 pKa = 3.61ICVLHH31 pKa = 6.92ANDD34 pKa = 4.38RR35 pKa = 11.84NVNTFGEE42 pKa = 4.27NFTNIFEE49 pKa = 4.87SYY51 pKa = 11.2NSVTLSFYY59 pKa = 11.1NYY61 pKa = 10.31DD62 pKa = 3.57SNNYY66 pKa = 9.78DD67 pKa = 3.8VIDD70 pKa = 3.82ILSKK74 pKa = 10.58KK75 pKa = 10.31DD76 pKa = 3.47YY77 pKa = 10.63SLCQILAIDD86 pKa = 4.34VIRR89 pKa = 11.84PEE91 pKa = 3.68MDD93 pKa = 4.33FITFLQSNNEE103 pKa = 3.75CSKK106 pKa = 10.69YY107 pKa = 10.16SRR109 pKa = 11.84QKK111 pKa = 9.79IHH113 pKa = 5.52YY114 pKa = 8.27QKK116 pKa = 10.11LTTGDD121 pKa = 2.86EE122 pKa = 3.97WFVYY126 pKa = 10.29SKK128 pKa = 11.0NLKK131 pKa = 9.78FCPLSDD137 pKa = 3.76NLIGLYY143 pKa = 10.43CDD145 pKa = 3.46TQVNGTYY152 pKa = 10.25FPLSNNEE159 pKa = 4.16RR160 pKa = 11.84YY161 pKa = 10.04DD162 pKa = 3.63VTDD165 pKa = 3.66LPEE168 pKa = 4.25FTEE171 pKa = 3.96MGYY174 pKa = 10.53VFYY177 pKa = 10.98SDD179 pKa = 4.25DD180 pKa = 4.04DD181 pKa = 4.4FYY183 pKa = 10.89ICKK186 pKa = 10.14RR187 pKa = 11.84VTEE190 pKa = 4.36EE191 pKa = 3.11KK192 pKa = 9.67WINYY196 pKa = 8.41HH197 pKa = 6.33LFYY200 pKa = 10.77RR201 pKa = 11.84EE202 pKa = 3.82YY203 pKa = 10.22SASGTVARR211 pKa = 11.84AISWDD216 pKa = 3.87NVWTGFKK223 pKa = 10.06TFAQVVYY230 pKa = 10.17KK231 pKa = 10.64ILDD234 pKa = 3.15IFFNNRR240 pKa = 11.84RR241 pKa = 11.84NFEE244 pKa = 3.84PRR246 pKa = 11.84AA247 pKa = 3.56
Molecular weight: 28.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6JPH2|Q6JPH2_9REOV Nonstructural protein 5 OS=Bovine group B rotavirus OX=35334 GN=NSP5 PE=4 SV=1
MM1 pKa = 7.53GSSQSSLQTQVHH13 pKa = 5.25STNIHH18 pKa = 5.07SQHH21 pKa = 6.8SSIHH25 pKa = 5.35LQGTSTATFTTHH37 pKa = 6.73QIIITAGAVLIALLLTSLVFSCICNCYY64 pKa = 10.24LYY66 pKa = 11.09SKK68 pKa = 9.9VKK70 pKa = 10.8NGFQTIPLQFKK81 pKa = 9.72RR82 pKa = 11.84KK83 pKa = 8.59EE84 pKa = 4.19RR85 pKa = 11.84FDD87 pKa = 3.71SSIPQPRR94 pKa = 11.84IQPTQFVV101 pKa = 3.57
MM1 pKa = 7.53GSSQSSLQTQVHH13 pKa = 5.25STNIHH18 pKa = 5.07SQHH21 pKa = 6.8SSIHH25 pKa = 5.35LQGTSTATFTTHH37 pKa = 6.73QIIITAGAVLIALLLTSLVFSCICNCYY64 pKa = 10.24LYY66 pKa = 11.09SKK68 pKa = 9.9VKK70 pKa = 10.8NGFQTIPLQFKK81 pKa = 9.72RR82 pKa = 11.84KK83 pKa = 8.59EE84 pKa = 4.19RR85 pKa = 11.84FDD87 pKa = 3.71SSIPQPRR94 pKa = 11.84IQPTQFVV101 pKa = 3.57
Molecular weight: 11.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4698 |
101 |
1158 |
469.8 |
53.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.13 ± 0.412 | 1.596 ± 0.571 |
5.811 ± 0.562 | 5.449 ± 0.358 |
4.47 ± 0.473 | 3.853 ± 0.449 |
1.724 ± 0.287 | 7.642 ± 0.314 |
6.301 ± 0.74 | 8.365 ± 0.556 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.15 | 6.003 ± 0.376 |
4.172 ± 0.349 | 3.917 ± 0.451 |
5.194 ± 0.399 | 8.472 ± 0.38 |
6.982 ± 0.498 | 6.599 ± 0.502 |
1.022 ± 0.158 | 3.959 ± 0.486 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
