
Actinomadura sp. LHW52907
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Actinomadura; unclassified Actinomadura
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
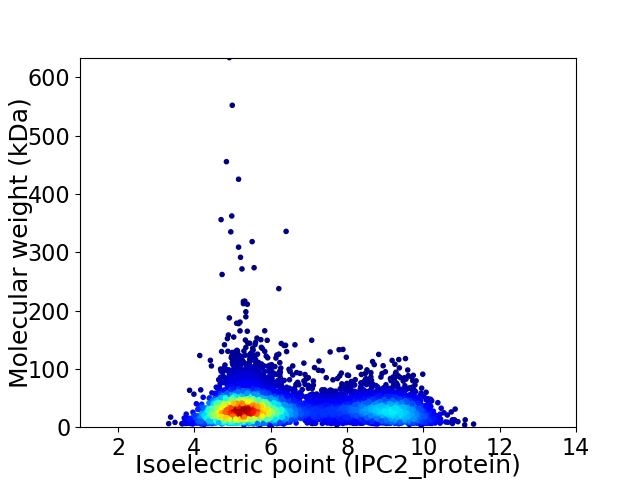
Virtual 2D-PAGE plot for 6669 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A372G8P6|A0A372G8P6_9ACTN Histidine kinase OS=Actinomadura sp. LHW52907 OX=2303421 GN=D0T12_30970 PE=4 SV=1
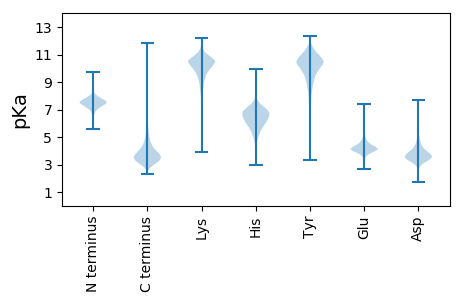
MM1 pKa = 7.24TSYY4 pKa = 10.71IVVFGLIVVVVLVVVLVALGLRR26 pKa = 11.84ASRR29 pKa = 11.84AGRR32 pKa = 11.84DD33 pKa = 3.44DD34 pKa = 5.19DD35 pKa = 5.62DD36 pKa = 4.1WMDD39 pKa = 5.69DD40 pKa = 3.5EE41 pKa = 4.53PQPRR45 pKa = 11.84GRR47 pKa = 11.84RR48 pKa = 11.84GAPQHH53 pKa = 6.35EE54 pKa = 4.86DD55 pKa = 2.81MGGPEE60 pKa = 4.31GYY62 pKa = 10.2GYY64 pKa = 11.01NDD66 pKa = 4.72GYY68 pKa = 10.53DD69 pKa = 2.95TGYY72 pKa = 11.2DD73 pKa = 2.99SGYY76 pKa = 10.54DD77 pKa = 3.22RR78 pKa = 11.84QGAPEE83 pKa = 4.22PAHH86 pKa = 6.61DD87 pKa = 3.89RR88 pKa = 11.84RR89 pKa = 11.84VAGGPLAAPTAQAAQGPPGPQVPQGPQAPPPPAPSGQASDD129 pKa = 4.46EE130 pKa = 4.3MADD133 pKa = 3.45DD134 pKa = 5.58DD135 pKa = 4.25YY136 pKa = 11.44WATITFDD143 pKa = 3.31KK144 pKa = 11.02PKK146 pKa = 10.48FPWQHH151 pKa = 5.87DD152 pKa = 3.68HH153 pKa = 7.38DD154 pKa = 4.54GEE156 pKa = 5.57RR157 pKa = 11.84GDD159 pKa = 4.18VDD161 pKa = 3.9PAADD165 pKa = 4.25PLNAQAPVEE174 pKa = 4.12QAPADD179 pKa = 3.61QPLPHH184 pKa = 6.38QPNLTQPVSMGADD197 pKa = 3.32GPTFGGMGAPGGQGGGTGPQQMAAPGGFQGGSPFANQDD235 pKa = 3.57PGSTALDD242 pKa = 5.0PIPADD247 pKa = 3.4LTGPGGPGPQGGPQPPYY264 pKa = 10.25GSGPGVPEE272 pKa = 3.52QPMYY276 pKa = 10.82GGQEE280 pKa = 4.13TYY282 pKa = 10.67GGQEE286 pKa = 3.76PQPAFGAADD295 pKa = 4.61FGQSDD300 pKa = 4.35QYY302 pKa = 11.72GGAPDD307 pKa = 4.27PVYY310 pKa = 10.95GDD312 pKa = 4.17PSPHH316 pKa = 5.84GTHH319 pKa = 6.19EE320 pKa = 4.15PSYY323 pKa = 11.3GSDD326 pKa = 3.4PLGGRR331 pKa = 11.84PAAAAGPPPAGRR343 pKa = 11.84QDD345 pKa = 3.74GYY347 pKa = 11.52DD348 pKa = 3.43FGLGSSGSGGGQPGGDD364 pKa = 3.43PRR366 pKa = 11.84VSDD369 pKa = 4.17PRR371 pKa = 11.84VSDD374 pKa = 3.62PRR376 pKa = 11.84VSDD379 pKa = 3.94PLGLPLGRR387 pKa = 11.84TDD389 pKa = 4.4EE390 pKa = 4.67PSAPPAPAAPAPPPPAAAAQGGAPGTDD417 pKa = 3.16TDD419 pKa = 3.73GHH421 pKa = 6.4KK422 pKa = 10.85LPTVDD427 pKa = 5.42EE428 pKa = 4.2LLQRR432 pKa = 11.84IQNDD436 pKa = 3.68RR437 pKa = 11.84QRR439 pKa = 11.84SSGSPSDD446 pKa = 3.68TGGGSYY452 pKa = 10.44GGSLNDD458 pKa = 4.35PLGDD462 pKa = 3.77PLGTGSFGTGSGTGTTGPWSTSGQTGGYY490 pKa = 10.33DD491 pKa = 2.96SGAPSGYY498 pKa = 10.2EE499 pKa = 3.64SGLGGTSGYY508 pKa = 10.93GQGQQSDD515 pKa = 5.0GYY517 pKa = 8.17PTAPAYY523 pKa = 10.42GDD525 pKa = 3.29SSRR528 pKa = 11.84YY529 pKa = 9.81DD530 pKa = 3.35DD531 pKa = 4.78PLNGTGRR538 pKa = 11.84EE539 pKa = 4.07PYY541 pKa = 9.3ATFDD545 pKa = 3.75GGSGTGTDD553 pKa = 3.68GQGVYY558 pKa = 11.09GDD560 pKa = 4.67FSGSSYY566 pKa = 11.52NGGADD571 pKa = 3.65PLSPPSDD578 pKa = 3.17QGASGGGYY586 pKa = 9.69GDD588 pKa = 4.34PGATQSYY595 pKa = 9.26GPGGYY600 pKa = 9.67GVPQPGGYY608 pKa = 8.26QQGHH612 pKa = 6.76PNDD615 pKa = 3.71TGPYY619 pKa = 9.71GSRR622 pKa = 11.84QPADD626 pKa = 2.9DD627 pKa = 3.59WEE629 pKa = 4.29NYY631 pKa = 8.27RR632 pKa = 11.84RR633 pKa = 3.78
MM1 pKa = 7.24TSYY4 pKa = 10.71IVVFGLIVVVVLVVVLVALGLRR26 pKa = 11.84ASRR29 pKa = 11.84AGRR32 pKa = 11.84DD33 pKa = 3.44DD34 pKa = 5.19DD35 pKa = 5.62DD36 pKa = 4.1WMDD39 pKa = 5.69DD40 pKa = 3.5EE41 pKa = 4.53PQPRR45 pKa = 11.84GRR47 pKa = 11.84RR48 pKa = 11.84GAPQHH53 pKa = 6.35EE54 pKa = 4.86DD55 pKa = 2.81MGGPEE60 pKa = 4.31GYY62 pKa = 10.2GYY64 pKa = 11.01NDD66 pKa = 4.72GYY68 pKa = 10.53DD69 pKa = 2.95TGYY72 pKa = 11.2DD73 pKa = 2.99SGYY76 pKa = 10.54DD77 pKa = 3.22RR78 pKa = 11.84QGAPEE83 pKa = 4.22PAHH86 pKa = 6.61DD87 pKa = 3.89RR88 pKa = 11.84RR89 pKa = 11.84VAGGPLAAPTAQAAQGPPGPQVPQGPQAPPPPAPSGQASDD129 pKa = 4.46EE130 pKa = 4.3MADD133 pKa = 3.45DD134 pKa = 5.58DD135 pKa = 4.25YY136 pKa = 11.44WATITFDD143 pKa = 3.31KK144 pKa = 11.02PKK146 pKa = 10.48FPWQHH151 pKa = 5.87DD152 pKa = 3.68HH153 pKa = 7.38DD154 pKa = 4.54GEE156 pKa = 5.57RR157 pKa = 11.84GDD159 pKa = 4.18VDD161 pKa = 3.9PAADD165 pKa = 4.25PLNAQAPVEE174 pKa = 4.12QAPADD179 pKa = 3.61QPLPHH184 pKa = 6.38QPNLTQPVSMGADD197 pKa = 3.32GPTFGGMGAPGGQGGGTGPQQMAAPGGFQGGSPFANQDD235 pKa = 3.57PGSTALDD242 pKa = 5.0PIPADD247 pKa = 3.4LTGPGGPGPQGGPQPPYY264 pKa = 10.25GSGPGVPEE272 pKa = 3.52QPMYY276 pKa = 10.82GGQEE280 pKa = 4.13TYY282 pKa = 10.67GGQEE286 pKa = 3.76PQPAFGAADD295 pKa = 4.61FGQSDD300 pKa = 4.35QYY302 pKa = 11.72GGAPDD307 pKa = 4.27PVYY310 pKa = 10.95GDD312 pKa = 4.17PSPHH316 pKa = 5.84GTHH319 pKa = 6.19EE320 pKa = 4.15PSYY323 pKa = 11.3GSDD326 pKa = 3.4PLGGRR331 pKa = 11.84PAAAAGPPPAGRR343 pKa = 11.84QDD345 pKa = 3.74GYY347 pKa = 11.52DD348 pKa = 3.43FGLGSSGSGGGQPGGDD364 pKa = 3.43PRR366 pKa = 11.84VSDD369 pKa = 4.17PRR371 pKa = 11.84VSDD374 pKa = 3.62PRR376 pKa = 11.84VSDD379 pKa = 3.94PLGLPLGRR387 pKa = 11.84TDD389 pKa = 4.4EE390 pKa = 4.67PSAPPAPAAPAPPPPAAAAQGGAPGTDD417 pKa = 3.16TDD419 pKa = 3.73GHH421 pKa = 6.4KK422 pKa = 10.85LPTVDD427 pKa = 5.42EE428 pKa = 4.2LLQRR432 pKa = 11.84IQNDD436 pKa = 3.68RR437 pKa = 11.84QRR439 pKa = 11.84SSGSPSDD446 pKa = 3.68TGGGSYY452 pKa = 10.44GGSLNDD458 pKa = 4.35PLGDD462 pKa = 3.77PLGTGSFGTGSGTGTTGPWSTSGQTGGYY490 pKa = 10.33DD491 pKa = 2.96SGAPSGYY498 pKa = 10.2EE499 pKa = 3.64SGLGGTSGYY508 pKa = 10.93GQGQQSDD515 pKa = 5.0GYY517 pKa = 8.17PTAPAYY523 pKa = 10.42GDD525 pKa = 3.29SSRR528 pKa = 11.84YY529 pKa = 9.81DD530 pKa = 3.35DD531 pKa = 4.78PLNGTGRR538 pKa = 11.84EE539 pKa = 4.07PYY541 pKa = 9.3ATFDD545 pKa = 3.75GGSGTGTDD553 pKa = 3.68GQGVYY558 pKa = 11.09GDD560 pKa = 4.67FSGSSYY566 pKa = 11.52NGGADD571 pKa = 3.65PLSPPSDD578 pKa = 3.17QGASGGGYY586 pKa = 9.69GDD588 pKa = 4.34PGATQSYY595 pKa = 9.26GPGGYY600 pKa = 9.67GVPQPGGYY608 pKa = 8.26QQGHH612 pKa = 6.76PNDD615 pKa = 3.71TGPYY619 pKa = 9.71GSRR622 pKa = 11.84QPADD626 pKa = 2.9DD627 pKa = 3.59WEE629 pKa = 4.29NYY631 pKa = 8.27RR632 pKa = 11.84RR633 pKa = 3.78
Molecular weight: 63.32 kDa
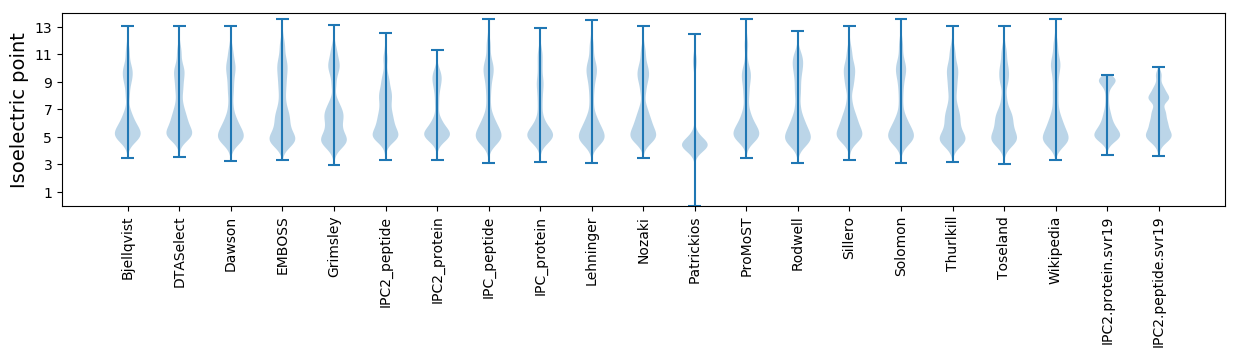
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A372GPP1|A0A372GPP1_9ACTN Slipin family protein OS=Actinomadura sp. LHW52907 OX=2303421 GN=D0T12_02165 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSNRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.31GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSNRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.31GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2203683 |
27 |
6004 |
330.4 |
35.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.064 ± 0.044 | 0.801 ± 0.009 |
6.349 ± 0.023 | 5.718 ± 0.025 |
2.768 ± 0.016 | 9.338 ± 0.031 |
2.253 ± 0.014 | 3.422 ± 0.023 |
1.978 ± 0.023 | 10.318 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.807 ± 0.014 | 1.767 ± 0.014 |
6.24 ± 0.028 | 2.443 ± 0.02 |
8.85 ± 0.035 | 4.75 ± 0.018 |
5.894 ± 0.031 | 8.758 ± 0.028 |
1.486 ± 0.012 | 1.998 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
