
Rothia mucilaginosa (strain DY-18) (Stomatococcus mucilaginosus)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Rothia; Rothia mucilaginosa
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
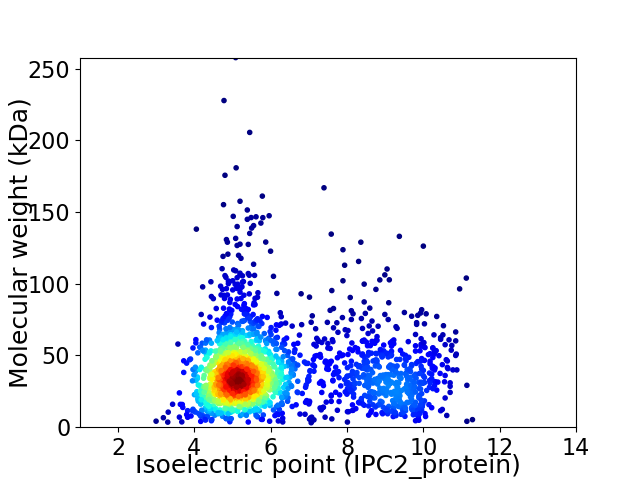
Virtual 2D-PAGE plot for 1991 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
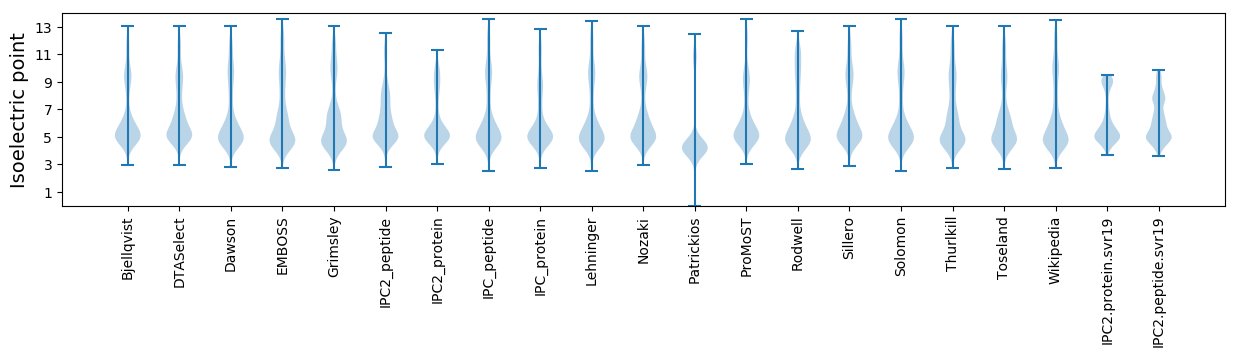
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2NTU7|D2NTU7_ROTMD ABC-type dipeptide/oligopeptide/nickel transport system permease component OS=Rothia mucilaginosa (strain DY-18) OX=680646 GN=RMDY18_12410 PE=4 SV=1
MM1 pKa = 7.23WPWYY5 pKa = 8.79TPSGNLARR13 pKa = 11.84IPRR16 pKa = 11.84FVILFSVLRR25 pKa = 11.84GGVLDD30 pKa = 4.66DD31 pKa = 5.45DD32 pKa = 4.63GVAVDD37 pKa = 4.45VEE39 pKa = 4.75SLTVPVSHH47 pKa = 7.28ASLLGVLDD55 pKa = 3.78SFEE58 pKa = 4.25EE59 pKa = 4.17KK60 pKa = 10.71LVGVFNGQGCEE71 pKa = 3.89GLLLTEE77 pKa = 4.83PLVEE81 pKa = 4.75GNDD84 pKa = 3.71GCGGFDD90 pKa = 3.35QVAVGTDD97 pKa = 3.18VEE99 pKa = 4.76FTLRR103 pKa = 11.84TSDD106 pKa = 3.93SVEE109 pKa = 3.9AEE111 pKa = 4.16CGKK114 pKa = 10.34EE115 pKa = 3.93CTRR118 pKa = 11.84SNVLGFVGTEE128 pKa = 3.71AQLTQGGEE136 pKa = 4.06AQNPGTDD143 pKa = 3.59DD144 pKa = 3.21TAACDD149 pKa = 3.55GTGFFEE155 pKa = 4.69RR156 pKa = 11.84GRR158 pKa = 11.84DD159 pKa = 3.42ATQAEE164 pKa = 4.94SGSGDD169 pKa = 3.21AGVSLGGLYY178 pKa = 10.27FLAACGNVLGAQLSFLVIDD197 pKa = 4.02EE198 pKa = 4.25RR199 pKa = 11.84VKK201 pKa = 10.42FLSVKK206 pKa = 10.3LYY208 pKa = 9.55TGGYY212 pKa = 9.44GINTGTATACLSVFSAGFLVCEE234 pKa = 4.73DD235 pKa = 4.43ADD237 pKa = 4.14DD238 pKa = 4.18FTDD241 pKa = 3.5EE242 pKa = 4.45GAAQAHH248 pKa = 6.0LVALASRR255 pKa = 11.84VPRR258 pKa = 11.84TEE260 pKa = 3.68EE261 pKa = 3.85EE262 pKa = 4.22RR263 pKa = 11.84CGGGVEE269 pKa = 4.31QCAHH273 pKa = 5.14VCGGAVAQCAVLVVFGDD290 pKa = 3.89GLTVDD295 pKa = 3.49VTGTEE300 pKa = 4.21VAIEE304 pKa = 3.97DD305 pKa = 3.84AVGEE309 pKa = 3.9FHH311 pKa = 7.42VGVFDD316 pKa = 3.64GTVFGLVGDD325 pKa = 4.9DD326 pKa = 3.81GAGNLDD332 pKa = 3.63DD333 pKa = 5.77CFVAVLAALFDD344 pKa = 4.25GGFCTVEE351 pKa = 3.78FRR353 pKa = 11.84LLAVNEE359 pKa = 4.19DD360 pKa = 3.75DD361 pKa = 5.54LLRR364 pKa = 11.84LALDD368 pKa = 3.7GLLGFVLVFRR378 pKa = 11.84LLYY381 pKa = 10.55DD382 pKa = 3.34VADD385 pKa = 4.84FVCEE389 pKa = 4.07DD390 pKa = 3.99RR391 pKa = 11.84LTVAPLTIDD400 pKa = 4.21KK401 pKa = 11.07LGGCQGCVGACCRR414 pKa = 11.84FLFSDD419 pKa = 4.11RR420 pKa = 11.84GDD422 pKa = 3.38NGAATEE428 pKa = 4.4LDD430 pKa = 3.58VLDD433 pKa = 4.49EE434 pKa = 4.37VGEE437 pKa = 4.35GDD439 pKa = 4.65DD440 pKa = 4.83GLGLVLGHH448 pKa = 6.7GCLLRR453 pKa = 5.67
MM1 pKa = 7.23WPWYY5 pKa = 8.79TPSGNLARR13 pKa = 11.84IPRR16 pKa = 11.84FVILFSVLRR25 pKa = 11.84GGVLDD30 pKa = 4.66DD31 pKa = 5.45DD32 pKa = 4.63GVAVDD37 pKa = 4.45VEE39 pKa = 4.75SLTVPVSHH47 pKa = 7.28ASLLGVLDD55 pKa = 3.78SFEE58 pKa = 4.25EE59 pKa = 4.17KK60 pKa = 10.71LVGVFNGQGCEE71 pKa = 3.89GLLLTEE77 pKa = 4.83PLVEE81 pKa = 4.75GNDD84 pKa = 3.71GCGGFDD90 pKa = 3.35QVAVGTDD97 pKa = 3.18VEE99 pKa = 4.76FTLRR103 pKa = 11.84TSDD106 pKa = 3.93SVEE109 pKa = 3.9AEE111 pKa = 4.16CGKK114 pKa = 10.34EE115 pKa = 3.93CTRR118 pKa = 11.84SNVLGFVGTEE128 pKa = 3.71AQLTQGGEE136 pKa = 4.06AQNPGTDD143 pKa = 3.59DD144 pKa = 3.21TAACDD149 pKa = 3.55GTGFFEE155 pKa = 4.69RR156 pKa = 11.84GRR158 pKa = 11.84DD159 pKa = 3.42ATQAEE164 pKa = 4.94SGSGDD169 pKa = 3.21AGVSLGGLYY178 pKa = 10.27FLAACGNVLGAQLSFLVIDD197 pKa = 4.02EE198 pKa = 4.25RR199 pKa = 11.84VKK201 pKa = 10.42FLSVKK206 pKa = 10.3LYY208 pKa = 9.55TGGYY212 pKa = 9.44GINTGTATACLSVFSAGFLVCEE234 pKa = 4.73DD235 pKa = 4.43ADD237 pKa = 4.14DD238 pKa = 4.18FTDD241 pKa = 3.5EE242 pKa = 4.45GAAQAHH248 pKa = 6.0LVALASRR255 pKa = 11.84VPRR258 pKa = 11.84TEE260 pKa = 3.68EE261 pKa = 3.85EE262 pKa = 4.22RR263 pKa = 11.84CGGGVEE269 pKa = 4.31QCAHH273 pKa = 5.14VCGGAVAQCAVLVVFGDD290 pKa = 3.89GLTVDD295 pKa = 3.49VTGTEE300 pKa = 4.21VAIEE304 pKa = 3.97DD305 pKa = 3.84AVGEE309 pKa = 3.9FHH311 pKa = 7.42VGVFDD316 pKa = 3.64GTVFGLVGDD325 pKa = 4.9DD326 pKa = 3.81GAGNLDD332 pKa = 3.63DD333 pKa = 5.77CFVAVLAALFDD344 pKa = 4.25GGFCTVEE351 pKa = 3.78FRR353 pKa = 11.84LLAVNEE359 pKa = 4.19DD360 pKa = 3.75DD361 pKa = 5.54LLRR364 pKa = 11.84LALDD368 pKa = 3.7GLLGFVLVFRR378 pKa = 11.84LLYY381 pKa = 10.55DD382 pKa = 3.34VADD385 pKa = 4.84FVCEE389 pKa = 4.07DD390 pKa = 3.99RR391 pKa = 11.84LTVAPLTIDD400 pKa = 4.21KK401 pKa = 11.07LGGCQGCVGACCRR414 pKa = 11.84FLFSDD419 pKa = 4.11RR420 pKa = 11.84GDD422 pKa = 3.38NGAATEE428 pKa = 4.4LDD430 pKa = 3.58VLDD433 pKa = 4.49EE434 pKa = 4.37VGEE437 pKa = 4.35GDD439 pKa = 4.65DD440 pKa = 4.83GLGLVLGHH448 pKa = 6.7GCLLRR453 pKa = 5.67
Molecular weight: 46.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2NQD8|D2NQD8_ROTMD EIIAB-Man OS=Rothia mucilaginosa (strain DY-18) OX=680646 GN=RMDY18_00320 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.51GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.51GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
757693 |
30 |
2358 |
380.6 |
41.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.593 ± 0.058 | 1.054 ± 0.032 |
5.012 ± 0.046 | 6.403 ± 0.06 |
3.055 ± 0.036 | 8.59 ± 0.072 |
2.647 ± 0.047 | 4.408 ± 0.05 |
3.141 ± 0.048 | 9.801 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.943 ± 0.029 | 3.337 ± 0.033 |
4.653 ± 0.05 | 3.841 ± 0.046 |
6.598 ± 0.06 | 6.348 ± 0.066 |
5.982 ± 0.048 | 8.207 ± 0.054 |
1.006 ± 0.022 | 2.379 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
