
Sandaracinus amylolyticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Sorangiineae; Sandaracinaceae; Sandaracinus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
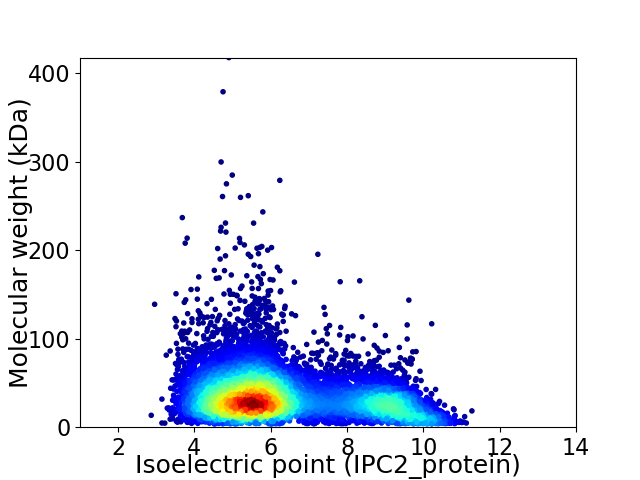
Virtual 2D-PAGE plot for 8941 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6YI92|A0A0F6YI92_9DELT Uncharacterized protein OS=Sandaracinus amylolyticus OX=927083 GN=DB32_003884 PE=4 SV=1
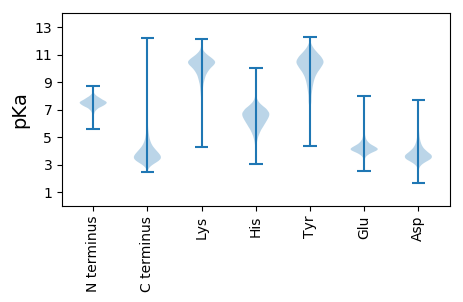
MM1 pKa = 7.3CRR3 pKa = 11.84WHH5 pKa = 7.34LDD7 pKa = 3.32HH8 pKa = 7.36ASCADD13 pKa = 3.98DD14 pKa = 3.98VFCDD18 pKa = 3.74GAEE21 pKa = 3.9RR22 pKa = 11.84CDD24 pKa = 3.84PVDD27 pKa = 3.12GCIEE31 pKa = 3.94GRR33 pKa = 11.84RR34 pKa = 11.84EE35 pKa = 3.93SCDD38 pKa = 3.6DD39 pKa = 3.8GDD41 pKa = 3.69VCTVDD46 pKa = 3.01RR47 pKa = 11.84CVEE50 pKa = 4.13SEE52 pKa = 4.2ASCTHH57 pKa = 7.29AARR60 pKa = 11.84DD61 pKa = 3.84LDD63 pKa = 4.33DD64 pKa = 6.24DD65 pKa = 5.02GDD67 pKa = 3.66VDD69 pKa = 4.02MFCDD73 pKa = 4.97GGNDD77 pKa = 4.5CDD79 pKa = 5.2DD80 pKa = 4.82ADD82 pKa = 4.08PTVSSGTDD90 pKa = 3.22EE91 pKa = 4.32RR92 pKa = 11.84CGNRR96 pKa = 11.84RR97 pKa = 11.84DD98 pKa = 4.05DD99 pKa = 4.8DD100 pKa = 4.36CDD102 pKa = 4.53GDD104 pKa = 3.86VDD106 pKa = 4.32EE107 pKa = 5.72DD108 pKa = 3.91ACATAPHH115 pKa = 6.78DD116 pKa = 3.99TCDD119 pKa = 4.21DD120 pKa = 4.32PLDD123 pKa = 3.85VSAGGSFVIEE133 pKa = 3.92ARR135 pKa = 11.84EE136 pKa = 3.89LRR138 pKa = 11.84GDD140 pKa = 3.69YY141 pKa = 10.22LGCGILEE148 pKa = 4.34CRR150 pKa = 11.84DD151 pKa = 3.4FVARR155 pKa = 11.84FTIDD159 pKa = 2.96EE160 pKa = 4.4AQDD163 pKa = 3.58VEE165 pKa = 5.65IIAADD170 pKa = 4.28PGDD173 pKa = 3.5DD174 pKa = 3.21WAYY177 pKa = 10.06TEE179 pKa = 3.95VALRR183 pKa = 11.84RR184 pKa = 11.84GCDD187 pKa = 3.38DD188 pKa = 3.41ATTEE192 pKa = 4.15IDD194 pKa = 4.27DD195 pKa = 3.89VTGFPATLRR204 pKa = 11.84YY205 pKa = 9.12RR206 pKa = 11.84ALEE209 pKa = 4.21PGTYY213 pKa = 9.4FVLAGAEE220 pKa = 4.17RR221 pKa = 11.84NFGSGATDD229 pKa = 3.31QLTLDD234 pKa = 3.82VTFSPDD240 pKa = 3.05TTPPTNEE247 pKa = 3.74TCASALDD254 pKa = 3.9VGAGGTFTADD264 pKa = 4.45FIDD267 pKa = 3.81TRR269 pKa = 11.84DD270 pKa = 4.1DD271 pKa = 3.58LALEE275 pKa = 4.64CGEE278 pKa = 4.9DD279 pKa = 3.47DD280 pKa = 3.91WPEE283 pKa = 3.52LVYY286 pKa = 10.69AITTSARR293 pKa = 11.84ADD295 pKa = 3.46LRR297 pKa = 11.84VTLDD301 pKa = 3.01GFARR305 pKa = 11.84LGLSVRR311 pKa = 11.84SACGDD316 pKa = 3.24ATSALGCVVLEE327 pKa = 3.99PHH329 pKa = 6.85EE330 pKa = 4.45DD331 pKa = 3.42RR332 pKa = 11.84VRR334 pKa = 11.84VFPDD338 pKa = 3.57VPAGTHH344 pKa = 5.16YY345 pKa = 10.44FVLEE349 pKa = 3.94LRR351 pKa = 11.84SIPEE355 pKa = 4.7AIAQITIEE363 pKa = 4.73ALPAAPP369 pKa = 4.86
MM1 pKa = 7.3CRR3 pKa = 11.84WHH5 pKa = 7.34LDD7 pKa = 3.32HH8 pKa = 7.36ASCADD13 pKa = 3.98DD14 pKa = 3.98VFCDD18 pKa = 3.74GAEE21 pKa = 3.9RR22 pKa = 11.84CDD24 pKa = 3.84PVDD27 pKa = 3.12GCIEE31 pKa = 3.94GRR33 pKa = 11.84RR34 pKa = 11.84EE35 pKa = 3.93SCDD38 pKa = 3.6DD39 pKa = 3.8GDD41 pKa = 3.69VCTVDD46 pKa = 3.01RR47 pKa = 11.84CVEE50 pKa = 4.13SEE52 pKa = 4.2ASCTHH57 pKa = 7.29AARR60 pKa = 11.84DD61 pKa = 3.84LDD63 pKa = 4.33DD64 pKa = 6.24DD65 pKa = 5.02GDD67 pKa = 3.66VDD69 pKa = 4.02MFCDD73 pKa = 4.97GGNDD77 pKa = 4.5CDD79 pKa = 5.2DD80 pKa = 4.82ADD82 pKa = 4.08PTVSSGTDD90 pKa = 3.22EE91 pKa = 4.32RR92 pKa = 11.84CGNRR96 pKa = 11.84RR97 pKa = 11.84DD98 pKa = 4.05DD99 pKa = 4.8DD100 pKa = 4.36CDD102 pKa = 4.53GDD104 pKa = 3.86VDD106 pKa = 4.32EE107 pKa = 5.72DD108 pKa = 3.91ACATAPHH115 pKa = 6.78DD116 pKa = 3.99TCDD119 pKa = 4.21DD120 pKa = 4.32PLDD123 pKa = 3.85VSAGGSFVIEE133 pKa = 3.92ARR135 pKa = 11.84EE136 pKa = 3.89LRR138 pKa = 11.84GDD140 pKa = 3.69YY141 pKa = 10.22LGCGILEE148 pKa = 4.34CRR150 pKa = 11.84DD151 pKa = 3.4FVARR155 pKa = 11.84FTIDD159 pKa = 2.96EE160 pKa = 4.4AQDD163 pKa = 3.58VEE165 pKa = 5.65IIAADD170 pKa = 4.28PGDD173 pKa = 3.5DD174 pKa = 3.21WAYY177 pKa = 10.06TEE179 pKa = 3.95VALRR183 pKa = 11.84RR184 pKa = 11.84GCDD187 pKa = 3.38DD188 pKa = 3.41ATTEE192 pKa = 4.15IDD194 pKa = 4.27DD195 pKa = 3.89VTGFPATLRR204 pKa = 11.84YY205 pKa = 9.12RR206 pKa = 11.84ALEE209 pKa = 4.21PGTYY213 pKa = 9.4FVLAGAEE220 pKa = 4.17RR221 pKa = 11.84NFGSGATDD229 pKa = 3.31QLTLDD234 pKa = 3.82VTFSPDD240 pKa = 3.05TTPPTNEE247 pKa = 3.74TCASALDD254 pKa = 3.9VGAGGTFTADD264 pKa = 4.45FIDD267 pKa = 3.81TRR269 pKa = 11.84DD270 pKa = 4.1DD271 pKa = 3.58LALEE275 pKa = 4.64CGEE278 pKa = 4.9DD279 pKa = 3.47DD280 pKa = 3.91WPEE283 pKa = 3.52LVYY286 pKa = 10.69AITTSARR293 pKa = 11.84ADD295 pKa = 3.46LRR297 pKa = 11.84VTLDD301 pKa = 3.01GFARR305 pKa = 11.84LGLSVRR311 pKa = 11.84SACGDD316 pKa = 3.24ATSALGCVVLEE327 pKa = 3.99PHH329 pKa = 6.85EE330 pKa = 4.45DD331 pKa = 3.42RR332 pKa = 11.84VRR334 pKa = 11.84VFPDD338 pKa = 3.57VPAGTHH344 pKa = 5.16YY345 pKa = 10.44FVLEE349 pKa = 3.94LRR351 pKa = 11.84SIPEE355 pKa = 4.7AIAQITIEE363 pKa = 4.73ALPAAPP369 pKa = 4.86
Molecular weight: 39.5 kDa
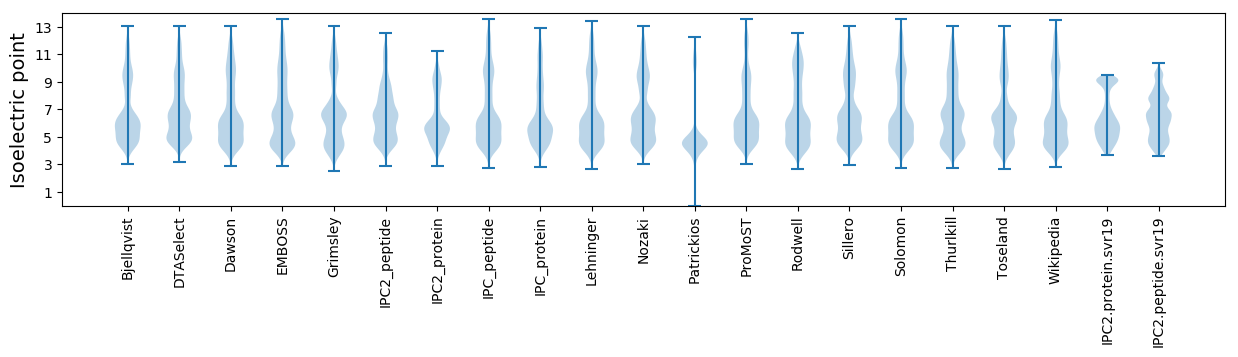
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6W5Z7|A0A0F6W5Z7_9DELT Histidine kinase OS=Sandaracinus amylolyticus OX=927083 GN=DB32_005441 PE=4 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84SARR5 pKa = 11.84GSARR9 pKa = 11.84RR10 pKa = 11.84PPRR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84ASTSTSPSAWRR28 pKa = 11.84AVPARR33 pKa = 11.84PRR35 pKa = 11.84AEE37 pKa = 3.83QRR39 pKa = 11.84PSVPTGRR46 pKa = 11.84HH47 pKa = 3.97AAIARR52 pKa = 11.84AAQARR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GPARR63 pKa = 11.84PIAGRR68 pKa = 11.84ARR70 pKa = 11.84AATAPVALRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84APPVRR86 pKa = 11.84PTARR90 pKa = 11.84PAPTASAPPASRR102 pKa = 11.84SRR104 pKa = 11.84RR105 pKa = 11.84APRR108 pKa = 11.84TAQSAEE114 pKa = 3.98TARR117 pKa = 11.84ARR119 pKa = 11.84PAPSRR124 pKa = 11.84PRR126 pKa = 11.84AARR129 pKa = 11.84RR130 pKa = 11.84TALRR134 pKa = 11.84AVKK137 pKa = 9.78PRR139 pKa = 11.84ARR141 pKa = 11.84AARR144 pKa = 11.84RR145 pKa = 11.84ASSARR150 pKa = 11.84STPCARR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84GSTARR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84PAARR169 pKa = 11.84RR170 pKa = 11.84SS171 pKa = 3.39
MM1 pKa = 7.64RR2 pKa = 11.84SARR5 pKa = 11.84GSARR9 pKa = 11.84RR10 pKa = 11.84PPRR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84ASTSTSPSAWRR28 pKa = 11.84AVPARR33 pKa = 11.84PRR35 pKa = 11.84AEE37 pKa = 3.83QRR39 pKa = 11.84PSVPTGRR46 pKa = 11.84HH47 pKa = 3.97AAIARR52 pKa = 11.84AAQARR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GPARR63 pKa = 11.84PIAGRR68 pKa = 11.84ARR70 pKa = 11.84AATAPVALRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84APPVRR86 pKa = 11.84PTARR90 pKa = 11.84PAPTASAPPASRR102 pKa = 11.84SRR104 pKa = 11.84RR105 pKa = 11.84APRR108 pKa = 11.84TAQSAEE114 pKa = 3.98TARR117 pKa = 11.84ARR119 pKa = 11.84PAPSRR124 pKa = 11.84PRR126 pKa = 11.84AARR129 pKa = 11.84RR130 pKa = 11.84TALRR134 pKa = 11.84AVKK137 pKa = 9.78PRR139 pKa = 11.84ARR141 pKa = 11.84AARR144 pKa = 11.84RR145 pKa = 11.84ASSARR150 pKa = 11.84STPCARR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84GSTARR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84PAARR169 pKa = 11.84RR170 pKa = 11.84SS171 pKa = 3.39
Molecular weight: 18.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3139928 |
37 |
3731 |
351.2 |
37.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.858 ± 0.046 | 1.331 ± 0.03 |
6.482 ± 0.029 | 6.542 ± 0.023 |
2.827 ± 0.015 | 8.793 ± 0.038 |
2.191 ± 0.012 | 3.889 ± 0.014 |
1.539 ± 0.022 | 9.604 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.842 ± 0.012 | 1.414 ± 0.014 |
5.796 ± 0.024 | 2.333 ± 0.015 |
9.455 ± 0.038 | 5.36 ± 0.02 |
5.297 ± 0.024 | 8.361 ± 0.025 |
1.369 ± 0.011 | 1.718 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
