
Desulfovibrio ferrophilus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
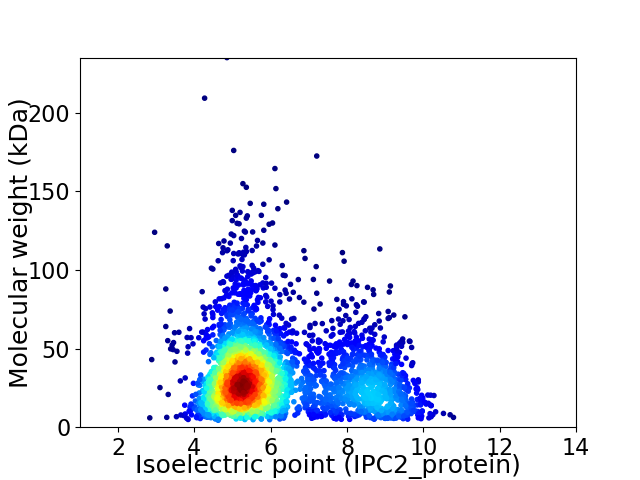
Virtual 2D-PAGE plot for 3428 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z6AWE5|A0A2Z6AWE5_9DELT Phosphotransferase system enzyme IIA component OS=Desulfovibrio ferrophilus OX=241368 GN=DFE_0851 PE=4 SV=1
MM1 pKa = 6.32YY2 pKa = 9.45TIEE5 pKa = 3.9ARR7 pKa = 11.84LYY9 pKa = 8.89QKK11 pKa = 10.52IMNDD15 pKa = 2.93QAAQTEE21 pKa = 4.39LLDD24 pKa = 4.51AGNDD28 pKa = 3.55QEE30 pKa = 4.38RR31 pKa = 11.84MEE33 pKa = 4.48VIVRR37 pKa = 11.84QGKK40 pKa = 7.97NHH42 pKa = 6.49GLHH45 pKa = 5.22VTPDD49 pKa = 3.44SVHH52 pKa = 6.2TFLKK56 pKa = 10.87DD57 pKa = 3.33KK58 pKa = 10.77MNTEE62 pKa = 4.23LSDD65 pKa = 3.88DD66 pKa = 3.83QLDD69 pKa = 3.77MVVGGKK75 pKa = 10.0GEE77 pKa = 3.98PASPNDD83 pKa = 3.89VIHH86 pKa = 6.92GNQFLDD92 pKa = 3.62LFGFTLDD99 pKa = 4.28DD100 pKa = 4.02NFDD103 pKa = 3.76GGEE106 pKa = 4.15GNDD109 pKa = 4.09YY110 pKa = 11.0ISGGWGNDD118 pKa = 3.02TLAGGTGNDD127 pKa = 3.44TLYY130 pKa = 11.14GGTGDD135 pKa = 3.64DD136 pKa = 3.52VMRR139 pKa = 11.84GGEE142 pKa = 3.96GHH144 pKa = 7.52DD145 pKa = 3.93YY146 pKa = 11.01MNGGQGNDD154 pKa = 3.39SMTGGQGNDD163 pKa = 3.05TMVGGDD169 pKa = 4.08GHH171 pKa = 8.2DD172 pKa = 3.4EE173 pKa = 4.32MYY175 pKa = 10.98GGTGNDD181 pKa = 3.18NMAGDD186 pKa = 4.27SGNDD190 pKa = 3.27TMYY193 pKa = 11.21GGANSDD199 pKa = 3.63TMDD202 pKa = 4.04GGSGNDD208 pKa = 2.84IMYY211 pKa = 10.32GGSGNDD217 pKa = 3.22SMEE220 pKa = 4.5GGLGDD225 pKa = 3.92DD226 pKa = 4.42LLRR229 pKa = 11.84GEE231 pKa = 5.07EE232 pKa = 4.39GDD234 pKa = 3.5DD235 pKa = 3.72TLRR238 pKa = 11.84GGYY241 pKa = 9.62GQDD244 pKa = 3.03TLYY247 pKa = 11.32GGAGNDD253 pKa = 3.95LLSGGLGSNNLYY265 pKa = 10.69GGEE268 pKa = 4.32GNDD271 pKa = 3.55TLLGGTASTYY281 pKa = 10.09MDD283 pKa = 4.22GGTGNDD289 pKa = 3.73LLVGHH294 pKa = 7.07TGSDD298 pKa = 3.1GMYY301 pKa = 10.48GGDD304 pKa = 4.21GHH306 pKa = 6.69DD307 pKa = 3.52TMYY310 pKa = 11.19GGAGNDD316 pKa = 3.5GMLGEE321 pKa = 5.2AGNDD325 pKa = 3.49YY326 pKa = 10.57MRR328 pKa = 11.84GDD330 pKa = 3.65SGNDD334 pKa = 3.12NMHH337 pKa = 6.66GGSGNDD343 pKa = 3.2TMFGGTGDD351 pKa = 4.29DD352 pKa = 3.93LLSGGEE358 pKa = 4.31GNDD361 pKa = 3.57VLHH364 pKa = 6.91GGAGTDD370 pKa = 3.45TLYY373 pKa = 11.34GGAGNDD379 pKa = 3.95TLWADD384 pKa = 3.52SGGGDD389 pKa = 3.9DD390 pKa = 4.61VLIGGSGADD399 pKa = 2.99VFVIGEE405 pKa = 3.97SDD407 pKa = 3.98GNVTIADD414 pKa = 4.39FNPEE418 pKa = 3.89EE419 pKa = 4.77DD420 pKa = 4.5KK421 pKa = 11.52LGLTNVPANGLVDD434 pKa = 3.28MDD436 pKa = 3.86VTNEE440 pKa = 3.83NGNTIIKK447 pKa = 10.61YY448 pKa = 10.46GDD450 pKa = 3.34TTVTLEE456 pKa = 3.8GVTLSGYY463 pKa = 7.22QVWDD467 pKa = 3.3ILDD470 pKa = 3.67TKK472 pKa = 11.11
MM1 pKa = 6.32YY2 pKa = 9.45TIEE5 pKa = 3.9ARR7 pKa = 11.84LYY9 pKa = 8.89QKK11 pKa = 10.52IMNDD15 pKa = 2.93QAAQTEE21 pKa = 4.39LLDD24 pKa = 4.51AGNDD28 pKa = 3.55QEE30 pKa = 4.38RR31 pKa = 11.84MEE33 pKa = 4.48VIVRR37 pKa = 11.84QGKK40 pKa = 7.97NHH42 pKa = 6.49GLHH45 pKa = 5.22VTPDD49 pKa = 3.44SVHH52 pKa = 6.2TFLKK56 pKa = 10.87DD57 pKa = 3.33KK58 pKa = 10.77MNTEE62 pKa = 4.23LSDD65 pKa = 3.88DD66 pKa = 3.83QLDD69 pKa = 3.77MVVGGKK75 pKa = 10.0GEE77 pKa = 3.98PASPNDD83 pKa = 3.89VIHH86 pKa = 6.92GNQFLDD92 pKa = 3.62LFGFTLDD99 pKa = 4.28DD100 pKa = 4.02NFDD103 pKa = 3.76GGEE106 pKa = 4.15GNDD109 pKa = 4.09YY110 pKa = 11.0ISGGWGNDD118 pKa = 3.02TLAGGTGNDD127 pKa = 3.44TLYY130 pKa = 11.14GGTGDD135 pKa = 3.64DD136 pKa = 3.52VMRR139 pKa = 11.84GGEE142 pKa = 3.96GHH144 pKa = 7.52DD145 pKa = 3.93YY146 pKa = 11.01MNGGQGNDD154 pKa = 3.39SMTGGQGNDD163 pKa = 3.05TMVGGDD169 pKa = 4.08GHH171 pKa = 8.2DD172 pKa = 3.4EE173 pKa = 4.32MYY175 pKa = 10.98GGTGNDD181 pKa = 3.18NMAGDD186 pKa = 4.27SGNDD190 pKa = 3.27TMYY193 pKa = 11.21GGANSDD199 pKa = 3.63TMDD202 pKa = 4.04GGSGNDD208 pKa = 2.84IMYY211 pKa = 10.32GGSGNDD217 pKa = 3.22SMEE220 pKa = 4.5GGLGDD225 pKa = 3.92DD226 pKa = 4.42LLRR229 pKa = 11.84GEE231 pKa = 5.07EE232 pKa = 4.39GDD234 pKa = 3.5DD235 pKa = 3.72TLRR238 pKa = 11.84GGYY241 pKa = 9.62GQDD244 pKa = 3.03TLYY247 pKa = 11.32GGAGNDD253 pKa = 3.95LLSGGLGSNNLYY265 pKa = 10.69GGEE268 pKa = 4.32GNDD271 pKa = 3.55TLLGGTASTYY281 pKa = 10.09MDD283 pKa = 4.22GGTGNDD289 pKa = 3.73LLVGHH294 pKa = 7.07TGSDD298 pKa = 3.1GMYY301 pKa = 10.48GGDD304 pKa = 4.21GHH306 pKa = 6.69DD307 pKa = 3.52TMYY310 pKa = 11.19GGAGNDD316 pKa = 3.5GMLGEE321 pKa = 5.2AGNDD325 pKa = 3.49YY326 pKa = 10.57MRR328 pKa = 11.84GDD330 pKa = 3.65SGNDD334 pKa = 3.12NMHH337 pKa = 6.66GGSGNDD343 pKa = 3.2TMFGGTGDD351 pKa = 4.29DD352 pKa = 3.93LLSGGEE358 pKa = 4.31GNDD361 pKa = 3.57VLHH364 pKa = 6.91GGAGTDD370 pKa = 3.45TLYY373 pKa = 11.34GGAGNDD379 pKa = 3.95TLWADD384 pKa = 3.52SGGGDD389 pKa = 3.9DD390 pKa = 4.61VLIGGSGADD399 pKa = 2.99VFVIGEE405 pKa = 3.97SDD407 pKa = 3.98GNVTIADD414 pKa = 4.39FNPEE418 pKa = 3.89EE419 pKa = 4.77DD420 pKa = 4.5KK421 pKa = 11.52LGLTNVPANGLVDD434 pKa = 3.28MDD436 pKa = 3.86VTNEE440 pKa = 3.83NGNTIIKK447 pKa = 10.61YY448 pKa = 10.46GDD450 pKa = 3.34TTVTLEE456 pKa = 3.8GVTLSGYY463 pKa = 7.22QVWDD467 pKa = 3.3ILDD470 pKa = 3.67TKK472 pKa = 11.11
Molecular weight: 48.24 kDa
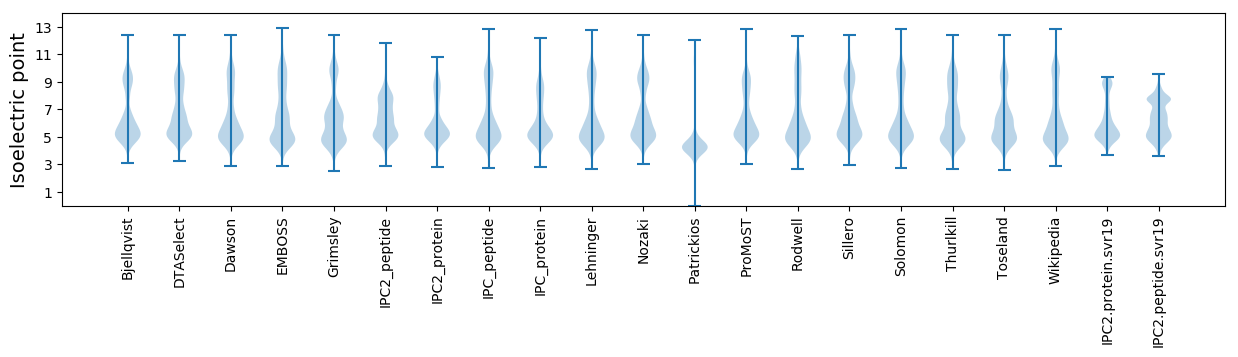
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6AZQ8|A0A2Z6AZQ8_9DELT Putative transcriptional regulator OS=Desulfovibrio ferrophilus OX=241368 GN=DFE_1954 PE=4 SV=1
MM1 pKa = 7.6PVLLTYY7 pKa = 10.8LPAILSMWLLGAHH20 pKa = 7.36ALRR23 pKa = 11.84GGQPGLMLAWALMPALFLLHH43 pKa = 6.95RR44 pKa = 11.84SWSHH48 pKa = 7.19RR49 pKa = 11.84IGQIALAAGVILWANTGSSLIQIRR73 pKa = 11.84IALDD77 pKa = 3.62QPWTRR82 pKa = 11.84LALILSGLILLAVLGIILLEE102 pKa = 4.17SQRR105 pKa = 11.84VATRR109 pKa = 11.84TQSDD113 pKa = 4.2TGPQWAPAWAFALCAGLLSTTQAKK137 pKa = 10.28VSFPILLPDD146 pKa = 3.8RR147 pKa = 11.84FLPGSGWLLILGLSIYY163 pKa = 10.42AAEE166 pKa = 4.24LTRR169 pKa = 11.84SFLMPGLKK177 pKa = 9.56PRR179 pKa = 11.84QWSTKK184 pKa = 10.02RR185 pKa = 11.84IRR187 pKa = 11.84LWGLFSAVFFVQLILGLAGAEE208 pKa = 3.76RR209 pKa = 11.84MLMTGQLHH217 pKa = 6.36LPIPALIMAGPLYY230 pKa = 10.29RR231 pKa = 11.84GEE233 pKa = 4.33GFFMLILFLSTVALVGPAWCSWLCYY258 pKa = 9.59IGAWDD263 pKa = 4.05GLSSRR268 pKa = 11.84TRR270 pKa = 11.84KK271 pKa = 7.59TPAPLPRR278 pKa = 11.84WRR280 pKa = 11.84QPVRR284 pKa = 11.84LGLFVLVAGTAWGLRR299 pKa = 11.84HH300 pKa = 6.14WGAPISVALALAAGFGLGGVAIMLTLSRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 9.94GVMVHH335 pKa = 6.05CTTFCPIGLLANILGKK351 pKa = 10.6LNPFRR356 pKa = 11.84VRR358 pKa = 11.84ITEE361 pKa = 4.0GCTGCGVCAAVCRR374 pKa = 11.84YY375 pKa = 9.25DD376 pKa = 4.23ALDD379 pKa = 3.58EE380 pKa = 4.04EE381 pKa = 5.49RR382 pKa = 11.84IALGKK387 pKa = 9.28PHH389 pKa = 7.5LSCTLCGDD397 pKa = 4.2CVGSCPHH404 pKa = 6.41TAMHH408 pKa = 6.23YY409 pKa = 8.89VFPGLSNANARR420 pKa = 11.84ALFVTMAVAAHH431 pKa = 6.21AVFLGVARR439 pKa = 11.84LL440 pKa = 3.49
MM1 pKa = 7.6PVLLTYY7 pKa = 10.8LPAILSMWLLGAHH20 pKa = 7.36ALRR23 pKa = 11.84GGQPGLMLAWALMPALFLLHH43 pKa = 6.95RR44 pKa = 11.84SWSHH48 pKa = 7.19RR49 pKa = 11.84IGQIALAAGVILWANTGSSLIQIRR73 pKa = 11.84IALDD77 pKa = 3.62QPWTRR82 pKa = 11.84LALILSGLILLAVLGIILLEE102 pKa = 4.17SQRR105 pKa = 11.84VATRR109 pKa = 11.84TQSDD113 pKa = 4.2TGPQWAPAWAFALCAGLLSTTQAKK137 pKa = 10.28VSFPILLPDD146 pKa = 3.8RR147 pKa = 11.84FLPGSGWLLILGLSIYY163 pKa = 10.42AAEE166 pKa = 4.24LTRR169 pKa = 11.84SFLMPGLKK177 pKa = 9.56PRR179 pKa = 11.84QWSTKK184 pKa = 10.02RR185 pKa = 11.84IRR187 pKa = 11.84LWGLFSAVFFVQLILGLAGAEE208 pKa = 3.76RR209 pKa = 11.84MLMTGQLHH217 pKa = 6.36LPIPALIMAGPLYY230 pKa = 10.29RR231 pKa = 11.84GEE233 pKa = 4.33GFFMLILFLSTVALVGPAWCSWLCYY258 pKa = 9.59IGAWDD263 pKa = 4.05GLSSRR268 pKa = 11.84TRR270 pKa = 11.84KK271 pKa = 7.59TPAPLPRR278 pKa = 11.84WRR280 pKa = 11.84QPVRR284 pKa = 11.84LGLFVLVAGTAWGLRR299 pKa = 11.84HH300 pKa = 6.14WGAPISVALALAAGFGLGGVAIMLTLSRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 9.94GVMVHH335 pKa = 6.05CTTFCPIGLLANILGKK351 pKa = 10.6LNPFRR356 pKa = 11.84VRR358 pKa = 11.84ITEE361 pKa = 4.0GCTGCGVCAAVCRR374 pKa = 11.84YY375 pKa = 9.25DD376 pKa = 4.23ALDD379 pKa = 3.58EE380 pKa = 4.04EE381 pKa = 5.49RR382 pKa = 11.84IALGKK387 pKa = 9.28PHH389 pKa = 7.5LSCTLCGDD397 pKa = 4.2CVGSCPHH404 pKa = 6.41TAMHH408 pKa = 6.23YY409 pKa = 8.89VFPGLSNANARR420 pKa = 11.84ALFVTMAVAAHH431 pKa = 6.21AVFLGVARR439 pKa = 11.84LL440 pKa = 3.49
Molecular weight: 47.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1088601 |
43 |
2103 |
317.6 |
34.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.55 ± 0.047 | 1.291 ± 0.021 |
5.739 ± 0.04 | 6.524 ± 0.04 |
3.938 ± 0.025 | 8.146 ± 0.056 |
2.113 ± 0.021 | 5.462 ± 0.034 |
4.619 ± 0.041 | 10.52 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.979 ± 0.021 | 3.131 ± 0.025 |
4.627 ± 0.034 | 3.424 ± 0.027 |
5.878 ± 0.034 | 5.751 ± 0.031 |
5.301 ± 0.026 | 7.223 ± 0.037 |
1.194 ± 0.016 | 2.591 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
