
Edaphobacter sp. 12200R-103
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Edaphobacter; unclassified Edaphobacter
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
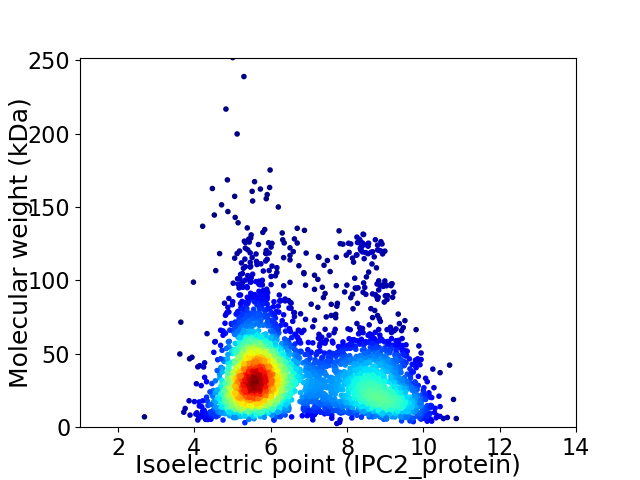
Virtual 2D-PAGE plot for 3688 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B9YNG7|A0A6B9YNG7_9BACT Flagellar basal body rod protein FlgB OS=Edaphobacter sp. 12200R-103 OX=2703788 GN=flgB PE=3 SV=1
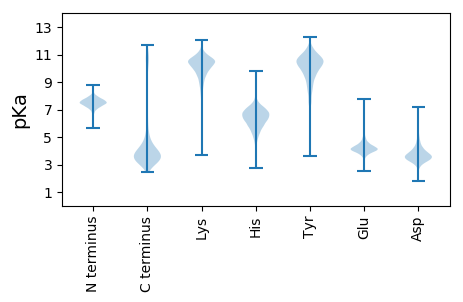
MM1 pKa = 8.06AIQADD6 pKa = 4.14FNGDD10 pKa = 3.07GRR12 pKa = 11.84PDD14 pKa = 3.62FFYY17 pKa = 11.04YY18 pKa = 10.65DD19 pKa = 3.39PLYY22 pKa = 10.75RR23 pKa = 11.84PNTSIRR29 pKa = 11.84TEE31 pKa = 3.8ILLSHH36 pKa = 7.2ADD38 pKa = 3.5GSYY41 pKa = 10.79SDD43 pKa = 4.61PLAVSLDD50 pKa = 3.68GTPQQCHH57 pKa = 6.6PYY59 pKa = 9.97DD60 pKa = 3.48INGDD64 pKa = 3.6HH65 pKa = 7.0FADD68 pKa = 4.42LVCISFASGSPALQVMYY85 pKa = 10.81SFIGNGDD92 pKa = 3.52GTFQPSIRR100 pKa = 11.84TDD102 pKa = 2.8ISINNDD108 pKa = 3.02FYY110 pKa = 11.79SSVGVTIAGDD120 pKa = 3.37INGDD124 pKa = 3.4GKK126 pKa = 11.37LDD128 pKa = 3.92VILTNLDD135 pKa = 3.79RR136 pKa = 11.84LNTIYY141 pKa = 10.77PMLGDD146 pKa = 3.8GAGHH150 pKa = 5.87FTLGKK155 pKa = 9.09PLNLRR160 pKa = 11.84YY161 pKa = 9.73YY162 pKa = 9.65SSGVRR167 pKa = 11.84LYY169 pKa = 11.25DD170 pKa = 3.48EE171 pKa = 4.57LHH173 pKa = 6.85DD174 pKa = 4.12VNGDD178 pKa = 3.44GKK180 pKa = 11.32LDD182 pKa = 3.94LLYY185 pKa = 10.62GQPLQVYY192 pKa = 9.14LGNGDD197 pKa = 3.68GTFTSGFIAGNYY209 pKa = 6.55QNCFFTDD216 pKa = 4.27FDD218 pKa = 4.27NDD220 pKa = 3.17QHH222 pKa = 8.41LDD224 pKa = 3.74AEE226 pKa = 4.83CVDD229 pKa = 3.58ISSNSNIILHH239 pKa = 6.31GNPDD243 pKa = 3.37GTFNTTPIATPDD255 pKa = 3.74FSNTTVLAIKK265 pKa = 10.03DD266 pKa = 3.92LNGDD270 pKa = 3.93GMPDD274 pKa = 3.96VISLGPKK281 pKa = 8.52GTIIYY286 pKa = 9.79LGKK289 pKa = 9.69PGLKK293 pKa = 9.34FAPPIAYY300 pKa = 9.72SFSAGYY306 pKa = 10.98SNFGVTDD313 pKa = 4.05PLIDD317 pKa = 5.13DD318 pKa = 4.11YY319 pKa = 11.97NGDD322 pKa = 4.35GILDD326 pKa = 4.39LATTAFDD333 pKa = 5.19GIYY336 pKa = 9.64IAYY339 pKa = 9.69GRR341 pKa = 11.84GDD343 pKa = 3.43GSFRR347 pKa = 11.84APRR350 pKa = 11.84PTEE353 pKa = 3.82SGTAIGGIAVGDD365 pKa = 3.94FDD367 pKa = 5.91EE368 pKa = 6.47DD369 pKa = 3.82GAPDD373 pKa = 4.44VITSGSPSLLLNHH386 pKa = 7.01GKK388 pKa = 10.83GDD390 pKa = 3.76GTFATSTPIQRR401 pKa = 11.84DD402 pKa = 3.26GLAANAGISSQVFQGDD418 pKa = 3.91FNGDD422 pKa = 2.97GHH424 pKa = 7.98LDD426 pKa = 3.41LVTNNSGSLAQIFFGKK442 pKa = 10.7GDD444 pKa = 3.75GTFSDD449 pKa = 4.53PVFASSTFNFIPFQGAVVADD469 pKa = 3.53INGDD473 pKa = 3.21GRR475 pKa = 11.84ADD477 pKa = 4.88LIAPNNSYY485 pKa = 8.33PTGDD489 pKa = 3.19VWVMLSNGDD498 pKa = 3.53GSFSAQSIATFNSALSVAAGDD519 pKa = 3.76VNGDD523 pKa = 3.34NKK525 pKa = 10.93LDD527 pKa = 3.89LVVSIAGGVQIFLGKK542 pKa = 10.66GDD544 pKa = 3.6GTFNQVVSAPAAPSTGNGTFSPGQAVFGDD573 pKa = 3.84FDD575 pKa = 5.49GDD577 pKa = 3.72GKK579 pKa = 11.07RR580 pKa = 11.84DD581 pKa = 3.57FVVTNNLSTYY591 pKa = 10.45YY592 pKa = 9.86LTIYY596 pKa = 9.65YY597 pKa = 10.61GNGDD601 pKa = 3.9GTFSAPSTIATGVSPTGLFLNLAAHH626 pKa = 7.02DD627 pKa = 4.54LNGDD631 pKa = 3.75GLDD634 pKa = 3.78DD635 pKa = 4.34LLYY638 pKa = 10.31SGSSIYY644 pKa = 10.21QVAEE648 pKa = 3.5ISILHH653 pKa = 6.51AKK655 pKa = 9.41PGRR658 pKa = 11.84TFDD661 pKa = 3.92AGMGLLAGFGTLTPSIADD679 pKa = 3.58FNKK682 pKa = 10.44DD683 pKa = 3.23GRR685 pKa = 11.84PDD687 pKa = 3.67LLFSNSYY694 pKa = 10.17ASPGIFTVLLNRR706 pKa = 11.84EE707 pKa = 4.39STGTTLTVDD716 pKa = 3.66HH717 pKa = 6.64TAIQYY722 pKa = 8.68GQGAGFTAQVTFSADD737 pKa = 3.31STSAPPPSATVILSGLPGSPVTLPIAFSSPGANAPFSGTARR778 pKa = 11.84YY779 pKa = 8.04EE780 pKa = 4.46AINLLPGTYY789 pKa = 10.29NVTARR794 pKa = 11.84LNASCCLTPSKK805 pKa = 10.81SAVTTLVVAPAATTTTLALTPVPPLTGQPATLRR838 pKa = 11.84VQVSSQTTVTSGTITFFDD856 pKa = 3.29QGTQIGTAPVASGAASFTTSPLSAGAHH883 pKa = 6.32SITAVFASDD892 pKa = 3.62QNFLTSSSQPLTVTVFDD909 pKa = 4.68RR910 pKa = 11.84GYY912 pKa = 10.79QLTAAQTNLSLGANGSTDD930 pKa = 4.33LVTLSPVQGFYY941 pKa = 11.49GKK943 pKa = 8.39VTLSWLGHH951 pKa = 6.24AGG953 pKa = 3.34
MM1 pKa = 8.06AIQADD6 pKa = 4.14FNGDD10 pKa = 3.07GRR12 pKa = 11.84PDD14 pKa = 3.62FFYY17 pKa = 11.04YY18 pKa = 10.65DD19 pKa = 3.39PLYY22 pKa = 10.75RR23 pKa = 11.84PNTSIRR29 pKa = 11.84TEE31 pKa = 3.8ILLSHH36 pKa = 7.2ADD38 pKa = 3.5GSYY41 pKa = 10.79SDD43 pKa = 4.61PLAVSLDD50 pKa = 3.68GTPQQCHH57 pKa = 6.6PYY59 pKa = 9.97DD60 pKa = 3.48INGDD64 pKa = 3.6HH65 pKa = 7.0FADD68 pKa = 4.42LVCISFASGSPALQVMYY85 pKa = 10.81SFIGNGDD92 pKa = 3.52GTFQPSIRR100 pKa = 11.84TDD102 pKa = 2.8ISINNDD108 pKa = 3.02FYY110 pKa = 11.79SSVGVTIAGDD120 pKa = 3.37INGDD124 pKa = 3.4GKK126 pKa = 11.37LDD128 pKa = 3.92VILTNLDD135 pKa = 3.79RR136 pKa = 11.84LNTIYY141 pKa = 10.77PMLGDD146 pKa = 3.8GAGHH150 pKa = 5.87FTLGKK155 pKa = 9.09PLNLRR160 pKa = 11.84YY161 pKa = 9.73YY162 pKa = 9.65SSGVRR167 pKa = 11.84LYY169 pKa = 11.25DD170 pKa = 3.48EE171 pKa = 4.57LHH173 pKa = 6.85DD174 pKa = 4.12VNGDD178 pKa = 3.44GKK180 pKa = 11.32LDD182 pKa = 3.94LLYY185 pKa = 10.62GQPLQVYY192 pKa = 9.14LGNGDD197 pKa = 3.68GTFTSGFIAGNYY209 pKa = 6.55QNCFFTDD216 pKa = 4.27FDD218 pKa = 4.27NDD220 pKa = 3.17QHH222 pKa = 8.41LDD224 pKa = 3.74AEE226 pKa = 4.83CVDD229 pKa = 3.58ISSNSNIILHH239 pKa = 6.31GNPDD243 pKa = 3.37GTFNTTPIATPDD255 pKa = 3.74FSNTTVLAIKK265 pKa = 10.03DD266 pKa = 3.92LNGDD270 pKa = 3.93GMPDD274 pKa = 3.96VISLGPKK281 pKa = 8.52GTIIYY286 pKa = 9.79LGKK289 pKa = 9.69PGLKK293 pKa = 9.34FAPPIAYY300 pKa = 9.72SFSAGYY306 pKa = 10.98SNFGVTDD313 pKa = 4.05PLIDD317 pKa = 5.13DD318 pKa = 4.11YY319 pKa = 11.97NGDD322 pKa = 4.35GILDD326 pKa = 4.39LATTAFDD333 pKa = 5.19GIYY336 pKa = 9.64IAYY339 pKa = 9.69GRR341 pKa = 11.84GDD343 pKa = 3.43GSFRR347 pKa = 11.84APRR350 pKa = 11.84PTEE353 pKa = 3.82SGTAIGGIAVGDD365 pKa = 3.94FDD367 pKa = 5.91EE368 pKa = 6.47DD369 pKa = 3.82GAPDD373 pKa = 4.44VITSGSPSLLLNHH386 pKa = 7.01GKK388 pKa = 10.83GDD390 pKa = 3.76GTFATSTPIQRR401 pKa = 11.84DD402 pKa = 3.26GLAANAGISSQVFQGDD418 pKa = 3.91FNGDD422 pKa = 2.97GHH424 pKa = 7.98LDD426 pKa = 3.41LVTNNSGSLAQIFFGKK442 pKa = 10.7GDD444 pKa = 3.75GTFSDD449 pKa = 4.53PVFASSTFNFIPFQGAVVADD469 pKa = 3.53INGDD473 pKa = 3.21GRR475 pKa = 11.84ADD477 pKa = 4.88LIAPNNSYY485 pKa = 8.33PTGDD489 pKa = 3.19VWVMLSNGDD498 pKa = 3.53GSFSAQSIATFNSALSVAAGDD519 pKa = 3.76VNGDD523 pKa = 3.34NKK525 pKa = 10.93LDD527 pKa = 3.89LVVSIAGGVQIFLGKK542 pKa = 10.66GDD544 pKa = 3.6GTFNQVVSAPAAPSTGNGTFSPGQAVFGDD573 pKa = 3.84FDD575 pKa = 5.49GDD577 pKa = 3.72GKK579 pKa = 11.07RR580 pKa = 11.84DD581 pKa = 3.57FVVTNNLSTYY591 pKa = 10.45YY592 pKa = 9.86LTIYY596 pKa = 9.65YY597 pKa = 10.61GNGDD601 pKa = 3.9GTFSAPSTIATGVSPTGLFLNLAAHH626 pKa = 7.02DD627 pKa = 4.54LNGDD631 pKa = 3.75GLDD634 pKa = 3.78DD635 pKa = 4.34LLYY638 pKa = 10.31SGSSIYY644 pKa = 10.21QVAEE648 pKa = 3.5ISILHH653 pKa = 6.51AKK655 pKa = 9.41PGRR658 pKa = 11.84TFDD661 pKa = 3.92AGMGLLAGFGTLTPSIADD679 pKa = 3.58FNKK682 pKa = 10.44DD683 pKa = 3.23GRR685 pKa = 11.84PDD687 pKa = 3.67LLFSNSYY694 pKa = 10.17ASPGIFTVLLNRR706 pKa = 11.84EE707 pKa = 4.39STGTTLTVDD716 pKa = 3.66HH717 pKa = 6.64TAIQYY722 pKa = 8.68GQGAGFTAQVTFSADD737 pKa = 3.31STSAPPPSATVILSGLPGSPVTLPIAFSSPGANAPFSGTARR778 pKa = 11.84YY779 pKa = 8.04EE780 pKa = 4.46AINLLPGTYY789 pKa = 10.29NVTARR794 pKa = 11.84LNASCCLTPSKK805 pKa = 10.81SAVTTLVVAPAATTTTLALTPVPPLTGQPATLRR838 pKa = 11.84VQVSSQTTVTSGTITFFDD856 pKa = 3.29QGTQIGTAPVASGAASFTTSPLSAGAHH883 pKa = 6.32SITAVFASDD892 pKa = 3.62QNFLTSSSQPLTVTVFDD909 pKa = 4.68RR910 pKa = 11.84GYY912 pKa = 10.79QLTAAQTNLSLGANGSTDD930 pKa = 4.33LVTLSPVQGFYY941 pKa = 11.49GKK943 pKa = 8.39VTLSWLGHH951 pKa = 6.24AGG953 pKa = 3.34
Molecular weight: 98.83 kDa
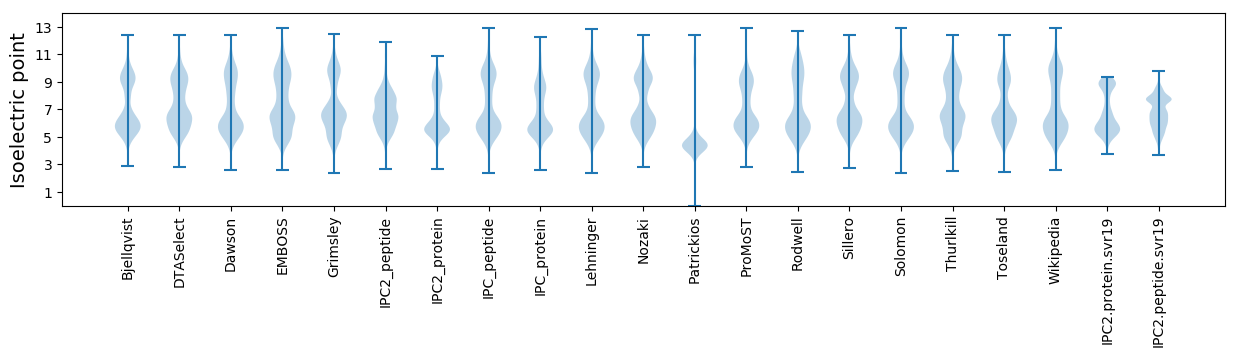
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9YS72|A0A6B9YS72_9BACT Patatin-like phospholipase family protein OS=Edaphobacter sp. 12200R-103 OX=2703788 GN=GWR55_12050 PE=4 SV=1
MM1 pKa = 6.98EE2 pKa = 4.71TVSLADD8 pKa = 2.92IDD10 pKa = 4.02KK11 pKa = 10.9FVVAKK16 pKa = 10.17RR17 pKa = 11.84AEE19 pKa = 4.17GCRR22 pKa = 11.84PRR24 pKa = 11.84GIANQCKK31 pKa = 9.72ALRR34 pKa = 11.84SFFRR38 pKa = 11.84FAEE41 pKa = 3.99MRR43 pKa = 11.84GWCVPNLALTIRR55 pKa = 11.84IPRR58 pKa = 11.84IPKK61 pKa = 9.71NEE63 pKa = 3.88PRR65 pKa = 11.84PTGPTWTQVRR75 pKa = 11.84QLLEE79 pKa = 4.03LTEE82 pKa = 4.82GNDD85 pKa = 3.57PEE87 pKa = 4.91HH88 pKa = 6.37IRR90 pKa = 11.84ARR92 pKa = 11.84ALLLLFTIYY101 pKa = 10.9GLRR104 pKa = 11.84ASEE107 pKa = 4.35VINLRR112 pKa = 11.84LEE114 pKa = 4.43DD115 pKa = 4.64FDD117 pKa = 3.76WQSEE121 pKa = 4.61VFTIRR126 pKa = 11.84RR127 pKa = 11.84AKK129 pKa = 10.56HH130 pKa = 6.17GGTQQYY136 pKa = 9.05PIQYY140 pKa = 8.96EE141 pKa = 3.98VGEE144 pKa = 4.99AILKK148 pKa = 7.24YY149 pKa = 10.27LRR151 pKa = 11.84YY152 pKa = 9.59VRR154 pKa = 11.84PRR156 pKa = 11.84VDD158 pKa = 3.33DD159 pKa = 3.48RR160 pKa = 11.84HH161 pKa = 6.19VFLGEE166 pKa = 3.8RR167 pKa = 11.84RR168 pKa = 11.84PWGPLQHH175 pKa = 4.97TTMWRR180 pKa = 11.84TVSRR184 pKa = 11.84RR185 pKa = 11.84LGALGIEE192 pKa = 4.67LRR194 pKa = 11.84HH195 pKa = 5.8QGPHH199 pKa = 6.56ALRR202 pKa = 11.84HH203 pKa = 5.05ACATRR208 pKa = 11.84LLQKK212 pKa = 10.65GSSLKK217 pKa = 10.59EE218 pKa = 3.43IADD221 pKa = 3.9FLGHH225 pKa = 7.08RR226 pKa = 11.84NTKK229 pKa = 10.29SVGIYY234 pKa = 9.85AKK236 pKa = 10.47CDD238 pKa = 2.83IAALRR243 pKa = 11.84KK244 pKa = 9.31VAAFRR249 pKa = 11.84LGGLRR254 pKa = 3.77
MM1 pKa = 6.98EE2 pKa = 4.71TVSLADD8 pKa = 2.92IDD10 pKa = 4.02KK11 pKa = 10.9FVVAKK16 pKa = 10.17RR17 pKa = 11.84AEE19 pKa = 4.17GCRR22 pKa = 11.84PRR24 pKa = 11.84GIANQCKK31 pKa = 9.72ALRR34 pKa = 11.84SFFRR38 pKa = 11.84FAEE41 pKa = 3.99MRR43 pKa = 11.84GWCVPNLALTIRR55 pKa = 11.84IPRR58 pKa = 11.84IPKK61 pKa = 9.71NEE63 pKa = 3.88PRR65 pKa = 11.84PTGPTWTQVRR75 pKa = 11.84QLLEE79 pKa = 4.03LTEE82 pKa = 4.82GNDD85 pKa = 3.57PEE87 pKa = 4.91HH88 pKa = 6.37IRR90 pKa = 11.84ARR92 pKa = 11.84ALLLLFTIYY101 pKa = 10.9GLRR104 pKa = 11.84ASEE107 pKa = 4.35VINLRR112 pKa = 11.84LEE114 pKa = 4.43DD115 pKa = 4.64FDD117 pKa = 3.76WQSEE121 pKa = 4.61VFTIRR126 pKa = 11.84RR127 pKa = 11.84AKK129 pKa = 10.56HH130 pKa = 6.17GGTQQYY136 pKa = 9.05PIQYY140 pKa = 8.96EE141 pKa = 3.98VGEE144 pKa = 4.99AILKK148 pKa = 7.24YY149 pKa = 10.27LRR151 pKa = 11.84YY152 pKa = 9.59VRR154 pKa = 11.84PRR156 pKa = 11.84VDD158 pKa = 3.33DD159 pKa = 3.48RR160 pKa = 11.84HH161 pKa = 6.19VFLGEE166 pKa = 3.8RR167 pKa = 11.84RR168 pKa = 11.84PWGPLQHH175 pKa = 4.97TTMWRR180 pKa = 11.84TVSRR184 pKa = 11.84RR185 pKa = 11.84LGALGIEE192 pKa = 4.67LRR194 pKa = 11.84HH195 pKa = 5.8QGPHH199 pKa = 6.56ALRR202 pKa = 11.84HH203 pKa = 5.05ACATRR208 pKa = 11.84LLQKK212 pKa = 10.65GSSLKK217 pKa = 10.59EE218 pKa = 3.43IADD221 pKa = 3.9FLGHH225 pKa = 7.08RR226 pKa = 11.84NTKK229 pKa = 10.29SVGIYY234 pKa = 9.85AKK236 pKa = 10.47CDD238 pKa = 2.83IAALRR243 pKa = 11.84KK244 pKa = 9.31VAAFRR249 pKa = 11.84LGGLRR254 pKa = 3.77
Molecular weight: 29.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1316478 |
24 |
2344 |
357.0 |
39.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.495 ± 0.05 | 0.851 ± 0.015 |
5.07 ± 0.024 | 5.504 ± 0.05 |
3.913 ± 0.026 | 8.031 ± 0.04 |
2.317 ± 0.019 | 5.256 ± 0.026 |
3.743 ± 0.034 | 9.802 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.319 ± 0.022 | 3.457 ± 0.044 |
5.332 ± 0.03 | 3.827 ± 0.026 |
6.339 ± 0.039 | 6.471 ± 0.036 |
5.962 ± 0.043 | 7.134 ± 0.032 |
1.383 ± 0.015 | 2.794 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
