
Staphylococcus massiliensis S46
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Staphylococcus; Staphylococcus massiliensis
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
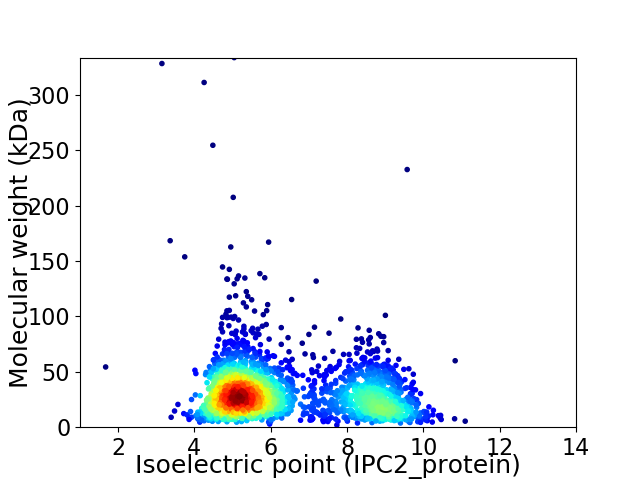
Virtual 2D-PAGE plot for 2323 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9B1Q3|K9B1Q3_9STAP 50S ribosomal subunit assembly factor BipA OS=Staphylococcus massiliensis S46 OX=1229783 GN=bipA PE=3 SV=1
MM1 pKa = 7.74EE2 pKa = 6.76DD3 pKa = 2.57IHH5 pKa = 9.03KK6 pKa = 10.63IIDD9 pKa = 4.78DD10 pKa = 3.97INLQNLKK17 pKa = 10.82GIDD20 pKa = 3.64SRR22 pKa = 11.84VEE24 pKa = 3.6NALTEE29 pKa = 4.22EE30 pKa = 4.14QDD32 pKa = 3.24EE33 pKa = 4.54GALFILGEE41 pKa = 4.07TLYY44 pKa = 10.7QYY46 pKa = 11.82GLIPQGLEE54 pKa = 3.68VFRR57 pKa = 11.84FLYY60 pKa = 10.46HH61 pKa = 6.96KK62 pKa = 10.37FPNEE66 pKa = 3.98PEE68 pKa = 3.78LLIYY72 pKa = 10.14FVEE75 pKa = 4.28GLIAEE80 pKa = 4.64GEE82 pKa = 4.02TDD84 pKa = 3.26EE85 pKa = 4.49ALEE88 pKa = 3.96YY89 pKa = 9.72LTEE92 pKa = 4.48VEE94 pKa = 4.53DD95 pKa = 4.25TPEE98 pKa = 4.52KK99 pKa = 11.02LMLEE103 pKa = 4.09ADD105 pKa = 5.01LYY107 pKa = 10.32QQLNMLEE114 pKa = 4.15VAIEE118 pKa = 4.13KK119 pKa = 10.46LNQAKK124 pKa = 9.62MIQPEE129 pKa = 3.93DD130 pKa = 4.27PIINFALAEE139 pKa = 3.99LFYY142 pKa = 11.38YY143 pKa = 10.49DD144 pKa = 4.25GQWLRR149 pKa = 11.84AVTEE153 pKa = 4.01YY154 pKa = 9.92DD155 pKa = 3.54TVLEE159 pKa = 4.46TGDD162 pKa = 3.57YY163 pKa = 9.37MINGVNLFSRR173 pKa = 11.84IADD176 pKa = 3.53CSLQSGNYY184 pKa = 9.51ADD186 pKa = 6.1AIQNYY191 pKa = 10.08DD192 pKa = 4.08EE193 pKa = 5.31ISDD196 pKa = 4.11SEE198 pKa = 4.45MTSEE202 pKa = 5.62DD203 pKa = 3.93YY204 pKa = 10.51FKK206 pKa = 11.15KK207 pKa = 10.62AISYY211 pKa = 7.54EE212 pKa = 4.05KK213 pKa = 10.86NEE215 pKa = 4.12RR216 pKa = 11.84TKK218 pKa = 10.94EE219 pKa = 4.12AISIMKK225 pKa = 10.15DD226 pKa = 3.43LLTKK230 pKa = 10.85DD231 pKa = 3.24PDD233 pKa = 4.61FIQGYY238 pKa = 9.23FYY240 pKa = 10.89LQQLYY245 pKa = 9.6EE246 pKa = 4.21LEE248 pKa = 4.15KK249 pKa = 10.31DD250 pKa = 3.7YY251 pKa = 11.29PSAIEE256 pKa = 4.27IGNEE260 pKa = 3.94GLKK263 pKa = 10.81LNQFYY268 pKa = 10.87KK269 pKa = 11.05EE270 pKa = 4.26LMCQTGSLEE279 pKa = 4.72IEE281 pKa = 4.87HH282 pKa = 7.27GDD284 pKa = 3.47QNDD287 pKa = 4.48GVDD290 pKa = 4.18HH291 pKa = 6.73LLEE294 pKa = 4.33SLEE297 pKa = 4.06VDD299 pKa = 3.92PSYY302 pKa = 11.15HH303 pKa = 6.18EE304 pKa = 4.16PVLILSEE311 pKa = 4.69FYY313 pKa = 10.64RR314 pKa = 11.84EE315 pKa = 4.11QDD317 pKa = 3.14DD318 pKa = 4.07HH319 pKa = 6.67EE320 pKa = 4.75ALIGLIKK327 pKa = 10.89YY328 pKa = 10.08VDD330 pKa = 4.32EE331 pKa = 4.74EE332 pKa = 4.58DD333 pKa = 3.57MDD335 pKa = 4.62PVFMWHH341 pKa = 6.75LAYY344 pKa = 10.07AYY346 pKa = 10.33GEE348 pKa = 4.13EE349 pKa = 4.36EE350 pKa = 3.88RR351 pKa = 11.84DD352 pKa = 3.25KK353 pKa = 11.48EE354 pKa = 4.44AMHH357 pKa = 7.03FYY359 pKa = 10.43QLAYY363 pKa = 8.84QTLNDD368 pKa = 3.72QAPFLSDD375 pKa = 3.34YY376 pKa = 10.69YY377 pKa = 10.94FYY379 pKa = 10.76LIEE382 pKa = 5.48IGHH385 pKa = 6.46IEE387 pKa = 3.99EE388 pKa = 5.16AKK390 pKa = 10.68PILQKK395 pKa = 10.87LKK397 pKa = 10.78EE398 pKa = 3.88IDD400 pKa = 3.45ATNEE404 pKa = 3.58TWFEE408 pKa = 4.06EE409 pKa = 4.15EE410 pKa = 6.06DD411 pKa = 3.52RR412 pKa = 11.84LQQ414 pKa = 4.27
MM1 pKa = 7.74EE2 pKa = 6.76DD3 pKa = 2.57IHH5 pKa = 9.03KK6 pKa = 10.63IIDD9 pKa = 4.78DD10 pKa = 3.97INLQNLKK17 pKa = 10.82GIDD20 pKa = 3.64SRR22 pKa = 11.84VEE24 pKa = 3.6NALTEE29 pKa = 4.22EE30 pKa = 4.14QDD32 pKa = 3.24EE33 pKa = 4.54GALFILGEE41 pKa = 4.07TLYY44 pKa = 10.7QYY46 pKa = 11.82GLIPQGLEE54 pKa = 3.68VFRR57 pKa = 11.84FLYY60 pKa = 10.46HH61 pKa = 6.96KK62 pKa = 10.37FPNEE66 pKa = 3.98PEE68 pKa = 3.78LLIYY72 pKa = 10.14FVEE75 pKa = 4.28GLIAEE80 pKa = 4.64GEE82 pKa = 4.02TDD84 pKa = 3.26EE85 pKa = 4.49ALEE88 pKa = 3.96YY89 pKa = 9.72LTEE92 pKa = 4.48VEE94 pKa = 4.53DD95 pKa = 4.25TPEE98 pKa = 4.52KK99 pKa = 11.02LMLEE103 pKa = 4.09ADD105 pKa = 5.01LYY107 pKa = 10.32QQLNMLEE114 pKa = 4.15VAIEE118 pKa = 4.13KK119 pKa = 10.46LNQAKK124 pKa = 9.62MIQPEE129 pKa = 3.93DD130 pKa = 4.27PIINFALAEE139 pKa = 3.99LFYY142 pKa = 11.38YY143 pKa = 10.49DD144 pKa = 4.25GQWLRR149 pKa = 11.84AVTEE153 pKa = 4.01YY154 pKa = 9.92DD155 pKa = 3.54TVLEE159 pKa = 4.46TGDD162 pKa = 3.57YY163 pKa = 9.37MINGVNLFSRR173 pKa = 11.84IADD176 pKa = 3.53CSLQSGNYY184 pKa = 9.51ADD186 pKa = 6.1AIQNYY191 pKa = 10.08DD192 pKa = 4.08EE193 pKa = 5.31ISDD196 pKa = 4.11SEE198 pKa = 4.45MTSEE202 pKa = 5.62DD203 pKa = 3.93YY204 pKa = 10.51FKK206 pKa = 11.15KK207 pKa = 10.62AISYY211 pKa = 7.54EE212 pKa = 4.05KK213 pKa = 10.86NEE215 pKa = 4.12RR216 pKa = 11.84TKK218 pKa = 10.94EE219 pKa = 4.12AISIMKK225 pKa = 10.15DD226 pKa = 3.43LLTKK230 pKa = 10.85DD231 pKa = 3.24PDD233 pKa = 4.61FIQGYY238 pKa = 9.23FYY240 pKa = 10.89LQQLYY245 pKa = 9.6EE246 pKa = 4.21LEE248 pKa = 4.15KK249 pKa = 10.31DD250 pKa = 3.7YY251 pKa = 11.29PSAIEE256 pKa = 4.27IGNEE260 pKa = 3.94GLKK263 pKa = 10.81LNQFYY268 pKa = 10.87KK269 pKa = 11.05EE270 pKa = 4.26LMCQTGSLEE279 pKa = 4.72IEE281 pKa = 4.87HH282 pKa = 7.27GDD284 pKa = 3.47QNDD287 pKa = 4.48GVDD290 pKa = 4.18HH291 pKa = 6.73LLEE294 pKa = 4.33SLEE297 pKa = 4.06VDD299 pKa = 3.92PSYY302 pKa = 11.15HH303 pKa = 6.18EE304 pKa = 4.16PVLILSEE311 pKa = 4.69FYY313 pKa = 10.64RR314 pKa = 11.84EE315 pKa = 4.11QDD317 pKa = 3.14DD318 pKa = 4.07HH319 pKa = 6.67EE320 pKa = 4.75ALIGLIKK327 pKa = 10.89YY328 pKa = 10.08VDD330 pKa = 4.32EE331 pKa = 4.74EE332 pKa = 4.58DD333 pKa = 3.57MDD335 pKa = 4.62PVFMWHH341 pKa = 6.75LAYY344 pKa = 10.07AYY346 pKa = 10.33GEE348 pKa = 4.13EE349 pKa = 4.36EE350 pKa = 3.88RR351 pKa = 11.84DD352 pKa = 3.25KK353 pKa = 11.48EE354 pKa = 4.44AMHH357 pKa = 7.03FYY359 pKa = 10.43QLAYY363 pKa = 8.84QTLNDD368 pKa = 3.72QAPFLSDD375 pKa = 3.34YY376 pKa = 10.69YY377 pKa = 10.94FYY379 pKa = 10.76LIEE382 pKa = 5.48IGHH385 pKa = 6.46IEE387 pKa = 3.99EE388 pKa = 5.16AKK390 pKa = 10.68PILQKK395 pKa = 10.87LKK397 pKa = 10.78EE398 pKa = 3.88IDD400 pKa = 3.45ATNEE404 pKa = 3.58TWFEE408 pKa = 4.06EE409 pKa = 4.15EE410 pKa = 6.06DD411 pKa = 3.52RR412 pKa = 11.84LQQ414 pKa = 4.27
Molecular weight: 48.43 kDa
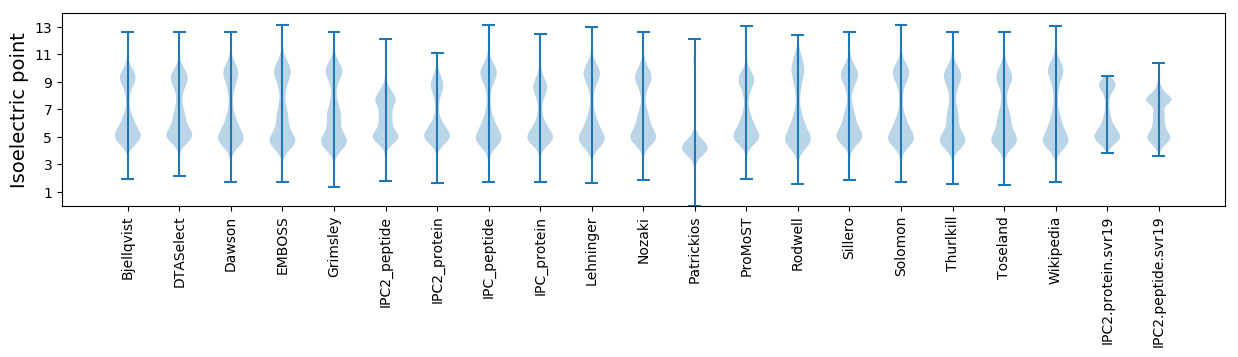
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9B413|K9B413_9STAP Competence protein F OS=Staphylococcus massiliensis S46 OX=1229783 GN=C273_04870 PE=4 SV=1
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.87VHH17 pKa = 5.77GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MSTKK26 pKa = 10.18NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.87VHH17 pKa = 5.77GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MSTKK26 pKa = 10.18NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
680527 |
18 |
3233 |
293.0 |
33.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.187 ± 0.05 | 0.639 ± 0.016 |
6.124 ± 0.071 | 7.032 ± 0.064 |
4.494 ± 0.042 | 6.281 ± 0.062 |
2.44 ± 0.03 | 8.258 ± 0.069 |
7.473 ± 0.064 | 9.317 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.745 ± 0.029 | 5.049 ± 0.056 |
3.328 ± 0.041 | 3.759 ± 0.042 |
3.759 ± 0.047 | 5.994 ± 0.05 |
5.723 ± 0.084 | 6.836 ± 0.047 |
0.723 ± 0.018 | 3.838 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
