
Salinibacterium sp. dk2585
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Salinibacterium; unclassified Salinibacterium
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
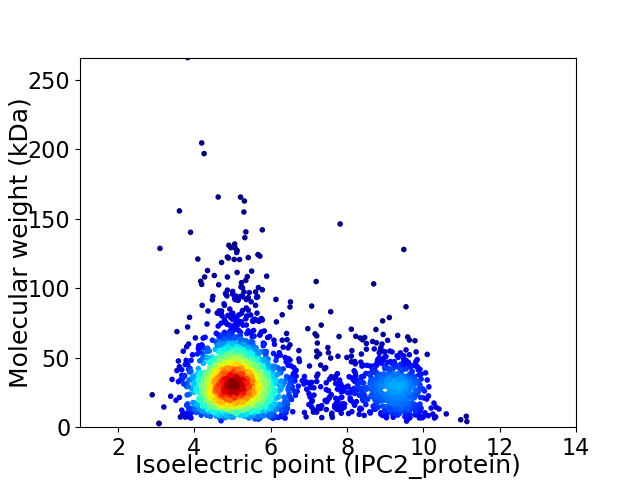
Virtual 2D-PAGE plot for 2617 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B9G5A0|A0A5B9G5A0_9MICO Uncharacterized protein OS=Salinibacterium sp. dk2585 OX=2603292 GN=FVA74_13220 PE=4 SV=1
MM1 pKa = 6.98HH2 pKa = 6.88TRR4 pKa = 11.84TRR6 pKa = 11.84RR7 pKa = 11.84QRR9 pKa = 11.84RR10 pKa = 11.84LGAVAGTALGAMLVTGCVASEE31 pKa = 4.01RR32 pKa = 11.84GEE34 pKa = 4.25DD35 pKa = 3.53SGDD38 pKa = 3.41AAGTFVFAASAEE50 pKa = 4.22PVTLDD55 pKa = 3.83PAFANDD61 pKa = 3.55GEE63 pKa = 4.63TFRR66 pKa = 11.84VARR69 pKa = 11.84QIFEE73 pKa = 3.94GLIGVEE79 pKa = 4.14QGSADD84 pKa = 4.05PAPLLAEE91 pKa = 4.07SWEE94 pKa = 4.31GSDD97 pKa = 5.84DD98 pKa = 3.74GLSYY102 pKa = 10.95TFDD105 pKa = 3.45LKK107 pKa = 11.17EE108 pKa = 3.97GVTFHH113 pKa = 7.87DD114 pKa = 3.63GTEE117 pKa = 4.32FNAEE121 pKa = 3.9AVCYY125 pKa = 10.81NFDD128 pKa = 3.21RR129 pKa = 11.84WHH131 pKa = 5.96NFEE134 pKa = 6.08GIAQSPSASYY144 pKa = 10.92YY145 pKa = 10.31YY146 pKa = 11.33NNLFKK151 pKa = 11.09GFAGDD156 pKa = 3.23EE157 pKa = 4.27GAVYY161 pKa = 9.18EE162 pKa = 4.44SCEE165 pKa = 3.86APSEE169 pKa = 4.19NEE171 pKa = 4.3AIVHH175 pKa = 6.03LTQPWAGFIAALSLPSFSMQSPTALQEE202 pKa = 3.9YY203 pKa = 9.62GADD206 pKa = 4.01DD207 pKa = 3.17IGGTEE212 pKa = 4.05EE213 pKa = 4.3APSISEE219 pKa = 4.21YY220 pKa = 10.93GSEE223 pKa = 4.46HH224 pKa = 6.0PTGTGPFTFSSWDD237 pKa = 3.1AGSQVEE243 pKa = 4.19LTAYY247 pKa = 10.33DD248 pKa = 4.69DD249 pKa = 3.73YY250 pKa = 11.13WGEE253 pKa = 4.05QGDD256 pKa = 3.9VTDD259 pKa = 4.14VIFRR263 pKa = 11.84VIDD266 pKa = 3.86DD267 pKa = 3.94PNARR271 pKa = 11.84RR272 pKa = 11.84QALEE276 pKa = 3.88AGTIDD281 pKa = 4.8GYY283 pKa = 11.41DD284 pKa = 3.47LVSPADD290 pKa = 3.64TAALEE295 pKa = 4.06EE296 pKa = 5.29DD297 pKa = 4.15GFQILARR304 pKa = 11.84DD305 pKa = 4.04PFTILYY311 pKa = 10.2LGINQAVPEE320 pKa = 4.29LQDD323 pKa = 3.1VRR325 pKa = 11.84VRR327 pKa = 11.84QAISMAIDD335 pKa = 3.34KK336 pKa = 9.24EE337 pKa = 4.22QLVNQTLPEE346 pKa = 4.23GTEE349 pKa = 3.95LATQFIPPVVSGYY362 pKa = 10.78NEE364 pKa = 4.65DD365 pKa = 3.64VTTYY369 pKa = 10.85DD370 pKa = 3.74YY371 pKa = 11.72DD372 pKa = 3.61VDD374 pKa = 3.7AAKK377 pKa = 10.84ALLAEE382 pKa = 4.54AGIPEE387 pKa = 4.18GWEE390 pKa = 3.62LTFNYY395 pKa = 6.61PTNVSRR401 pKa = 11.84PYY403 pKa = 9.52MPNPEE408 pKa = 4.49EE409 pKa = 4.14IFTVLSTQLSEE420 pKa = 3.85IGITVVPQANEE431 pKa = 3.26WGEE434 pKa = 4.1YY435 pKa = 9.71LDD437 pKa = 4.58IMQGSSDD444 pKa = 3.79HH445 pKa = 6.87GIHH448 pKa = 6.59LLGWTGDD455 pKa = 3.85YY456 pKa = 11.17NDD458 pKa = 3.83TDD460 pKa = 3.59NFVGVFFGQPSMEE473 pKa = 4.23FGFDD477 pKa = 3.3NPEE480 pKa = 3.99LFNALTEE487 pKa = 4.28ARR489 pKa = 11.84GTATIEE495 pKa = 4.09EE496 pKa = 4.23QLPLYY501 pKa = 10.56EE502 pKa = 6.2DD503 pKa = 3.77INQMIAEE510 pKa = 4.71YY511 pKa = 10.32IPAVPLAHH519 pKa = 7.21PAPSLAFSEE528 pKa = 4.98RR529 pKa = 11.84VGSYY533 pKa = 9.65PVSPVNDD540 pKa = 3.14EE541 pKa = 4.23VYY543 pKa = 11.31NLVEE547 pKa = 4.82LNDD550 pKa = 3.62
MM1 pKa = 6.98HH2 pKa = 6.88TRR4 pKa = 11.84TRR6 pKa = 11.84RR7 pKa = 11.84QRR9 pKa = 11.84RR10 pKa = 11.84LGAVAGTALGAMLVTGCVASEE31 pKa = 4.01RR32 pKa = 11.84GEE34 pKa = 4.25DD35 pKa = 3.53SGDD38 pKa = 3.41AAGTFVFAASAEE50 pKa = 4.22PVTLDD55 pKa = 3.83PAFANDD61 pKa = 3.55GEE63 pKa = 4.63TFRR66 pKa = 11.84VARR69 pKa = 11.84QIFEE73 pKa = 3.94GLIGVEE79 pKa = 4.14QGSADD84 pKa = 4.05PAPLLAEE91 pKa = 4.07SWEE94 pKa = 4.31GSDD97 pKa = 5.84DD98 pKa = 3.74GLSYY102 pKa = 10.95TFDD105 pKa = 3.45LKK107 pKa = 11.17EE108 pKa = 3.97GVTFHH113 pKa = 7.87DD114 pKa = 3.63GTEE117 pKa = 4.32FNAEE121 pKa = 3.9AVCYY125 pKa = 10.81NFDD128 pKa = 3.21RR129 pKa = 11.84WHH131 pKa = 5.96NFEE134 pKa = 6.08GIAQSPSASYY144 pKa = 10.92YY145 pKa = 10.31YY146 pKa = 11.33NNLFKK151 pKa = 11.09GFAGDD156 pKa = 3.23EE157 pKa = 4.27GAVYY161 pKa = 9.18EE162 pKa = 4.44SCEE165 pKa = 3.86APSEE169 pKa = 4.19NEE171 pKa = 4.3AIVHH175 pKa = 6.03LTQPWAGFIAALSLPSFSMQSPTALQEE202 pKa = 3.9YY203 pKa = 9.62GADD206 pKa = 4.01DD207 pKa = 3.17IGGTEE212 pKa = 4.05EE213 pKa = 4.3APSISEE219 pKa = 4.21YY220 pKa = 10.93GSEE223 pKa = 4.46HH224 pKa = 6.0PTGTGPFTFSSWDD237 pKa = 3.1AGSQVEE243 pKa = 4.19LTAYY247 pKa = 10.33DD248 pKa = 4.69DD249 pKa = 3.73YY250 pKa = 11.13WGEE253 pKa = 4.05QGDD256 pKa = 3.9VTDD259 pKa = 4.14VIFRR263 pKa = 11.84VIDD266 pKa = 3.86DD267 pKa = 3.94PNARR271 pKa = 11.84RR272 pKa = 11.84QALEE276 pKa = 3.88AGTIDD281 pKa = 4.8GYY283 pKa = 11.41DD284 pKa = 3.47LVSPADD290 pKa = 3.64TAALEE295 pKa = 4.06EE296 pKa = 5.29DD297 pKa = 4.15GFQILARR304 pKa = 11.84DD305 pKa = 4.04PFTILYY311 pKa = 10.2LGINQAVPEE320 pKa = 4.29LQDD323 pKa = 3.1VRR325 pKa = 11.84VRR327 pKa = 11.84QAISMAIDD335 pKa = 3.34KK336 pKa = 9.24EE337 pKa = 4.22QLVNQTLPEE346 pKa = 4.23GTEE349 pKa = 3.95LATQFIPPVVSGYY362 pKa = 10.78NEE364 pKa = 4.65DD365 pKa = 3.64VTTYY369 pKa = 10.85DD370 pKa = 3.74YY371 pKa = 11.72DD372 pKa = 3.61VDD374 pKa = 3.7AAKK377 pKa = 10.84ALLAEE382 pKa = 4.54AGIPEE387 pKa = 4.18GWEE390 pKa = 3.62LTFNYY395 pKa = 6.61PTNVSRR401 pKa = 11.84PYY403 pKa = 9.52MPNPEE408 pKa = 4.49EE409 pKa = 4.14IFTVLSTQLSEE420 pKa = 3.85IGITVVPQANEE431 pKa = 3.26WGEE434 pKa = 4.1YY435 pKa = 9.71LDD437 pKa = 4.58IMQGSSDD444 pKa = 3.79HH445 pKa = 6.87GIHH448 pKa = 6.59LLGWTGDD455 pKa = 3.85YY456 pKa = 11.17NDD458 pKa = 3.83TDD460 pKa = 3.59NFVGVFFGQPSMEE473 pKa = 4.23FGFDD477 pKa = 3.3NPEE480 pKa = 3.99LFNALTEE487 pKa = 4.28ARR489 pKa = 11.84GTATIEE495 pKa = 4.09EE496 pKa = 4.23QLPLYY501 pKa = 10.56EE502 pKa = 6.2DD503 pKa = 3.77INQMIAEE510 pKa = 4.71YY511 pKa = 10.32IPAVPLAHH519 pKa = 7.21PAPSLAFSEE528 pKa = 4.98RR529 pKa = 11.84VGSYY533 pKa = 9.65PVSPVNDD540 pKa = 3.14EE541 pKa = 4.23VYY543 pKa = 11.31NLVEE547 pKa = 4.82LNDD550 pKa = 3.62
Molecular weight: 59.87 kDa
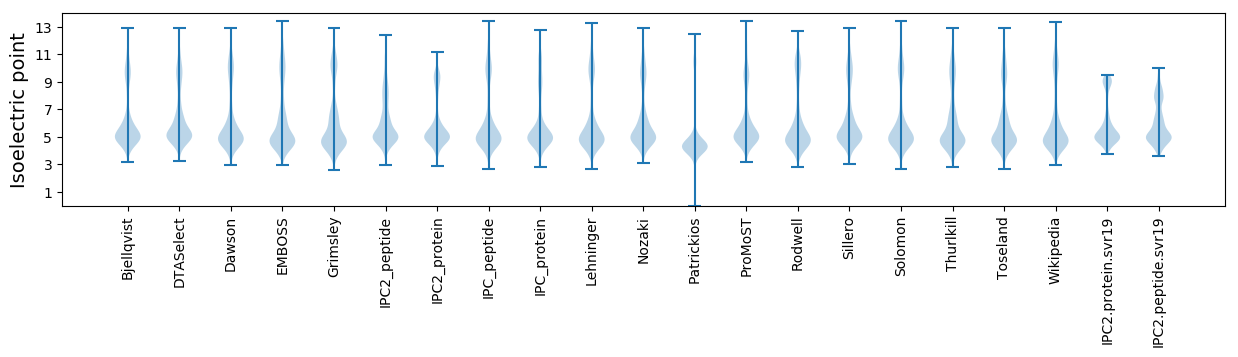
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9G5D1|A0A5B9G5D1_9MICO SDR family oxidoreductase OS=Salinibacterium sp. dk2585 OX=2603292 GN=FVA74_05045 PE=4 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84AITFATGTSVAGRR15 pKa = 11.84LTLRR19 pKa = 11.84APRR22 pKa = 11.84TIVVTQRR29 pKa = 11.84PSVIASTSGVASPGGVAPRR48 pKa = 11.84GLAATRR54 pKa = 11.84FPFRR58 pKa = 11.84GAAVVIASAVRR69 pKa = 11.84ALTWSAGTII78 pKa = 3.63
MM1 pKa = 7.68RR2 pKa = 11.84AITFATGTSVAGRR15 pKa = 11.84LTLRR19 pKa = 11.84APRR22 pKa = 11.84TIVVTQRR29 pKa = 11.84PSVIASTSGVASPGGVAPRR48 pKa = 11.84GLAATRR54 pKa = 11.84FPFRR58 pKa = 11.84GAAVVIASAVRR69 pKa = 11.84ALTWSAGTII78 pKa = 3.63
Molecular weight: 7.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
860142 |
24 |
2600 |
328.7 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.903 ± 0.063 | 0.503 ± 0.011 |
5.809 ± 0.039 | 6.207 ± 0.055 |
3.169 ± 0.03 | 8.822 ± 0.043 |
2.005 ± 0.021 | 4.758 ± 0.042 |
2.21 ± 0.035 | 10.147 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.954 ± 0.021 | 2.145 ± 0.024 |
5.204 ± 0.032 | 2.826 ± 0.024 |
7.071 ± 0.059 | 6.026 ± 0.035 |
5.916 ± 0.054 | 8.929 ± 0.047 |
1.406 ± 0.022 | 1.993 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
