
Protaetiibacter intestinalis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Protaetiibacter
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
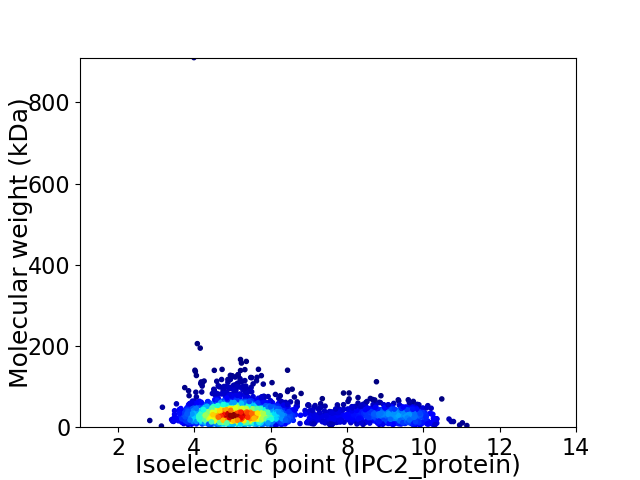
Virtual 2D-PAGE plot for 2836 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
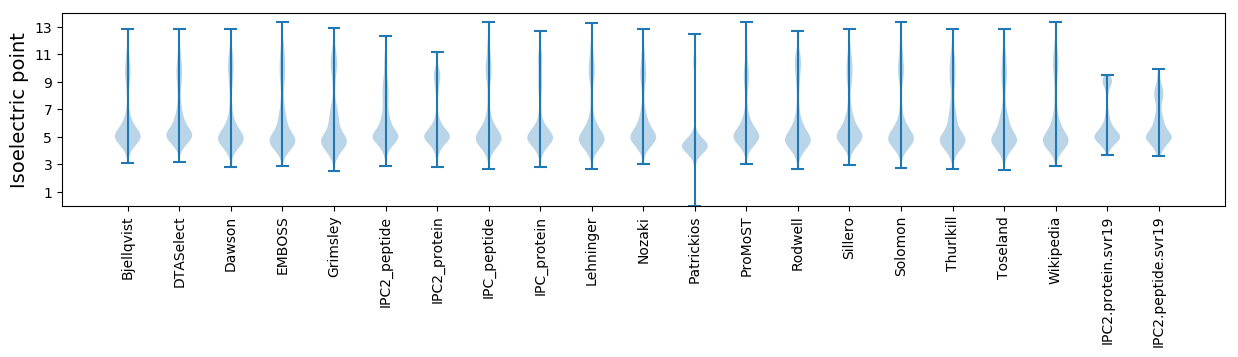
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A387B928|A0A387B928_9MICO Uncharacterized protein OS=Protaetiibacter intestinalis OX=2419774 GN=D7I47_11720 PE=4 SV=1
MM1 pKa = 7.33HH2 pKa = 7.23AHH4 pKa = 7.56SIRR7 pKa = 11.84TGLGIGAVTAVALALVGIPSAIAAADD33 pKa = 4.05GPVEE37 pKa = 4.04AGIFVDD43 pKa = 5.99RR44 pKa = 11.84IDD46 pKa = 4.1GLGDD50 pKa = 3.53DD51 pKa = 6.05FINGVDD57 pKa = 3.55VSSILSEE64 pKa = 4.15EE65 pKa = 4.19EE66 pKa = 4.02SGVVYY71 pKa = 10.86YY72 pKa = 9.45DD73 pKa = 3.91TEE75 pKa = 4.06GHH77 pKa = 5.8QADD80 pKa = 4.62LFDD83 pKa = 3.9VLEE86 pKa = 4.86DD87 pKa = 3.53ADD89 pKa = 3.7INYY92 pKa = 9.17VRR94 pKa = 11.84VRR96 pKa = 11.84VWDD99 pKa = 3.94DD100 pKa = 3.55PYY102 pKa = 11.28DD103 pKa = 4.16AEE105 pKa = 4.58GNGYY109 pKa = 10.66GGGTTDD115 pKa = 4.07LARR118 pKa = 11.84AVEE121 pKa = 3.99IGEE124 pKa = 4.14RR125 pKa = 11.84ATAHH129 pKa = 5.76GMRR132 pKa = 11.84VLVDD136 pKa = 4.63FHH138 pKa = 8.88YY139 pKa = 11.36SDD141 pKa = 4.96FWADD145 pKa = 3.24PAKK148 pKa = 10.54QHH150 pKa = 6.15VPKK153 pKa = 10.36AWEE156 pKa = 5.08GYY158 pKa = 5.39TTEE161 pKa = 4.21QLVTALGAYY170 pKa = 8.85TEE172 pKa = 4.49ASLDD176 pKa = 3.51AFADD180 pKa = 3.59AGVDD184 pKa = 3.26VGMVQIGNEE193 pKa = 4.27TNNTIAGVADD203 pKa = 3.82WDD205 pKa = 3.87GKK207 pKa = 11.07AAMFSAGSAAVRR219 pKa = 11.84EE220 pKa = 4.41VFPDD224 pKa = 3.43ALVALHH230 pKa = 6.12FTNPEE235 pKa = 3.73TPNRR239 pKa = 11.84YY240 pKa = 7.64TGYY243 pKa = 10.17AAQLAAHH250 pKa = 6.74DD251 pKa = 3.65VDD253 pKa = 4.06YY254 pKa = 11.5DD255 pKa = 3.9VFASSYY261 pKa = 8.41YY262 pKa = 10.06NFWHH266 pKa = 7.01GSPEE270 pKa = 3.94NLTSVLSTIADD281 pKa = 3.81SYY283 pKa = 10.9DD284 pKa = 3.37KK285 pKa = 10.99KK286 pKa = 11.66VMVAEE291 pKa = 4.32TSYY294 pKa = 11.72VYY296 pKa = 10.66TFDD299 pKa = 6.26DD300 pKa = 4.01GDD302 pKa = 3.34GWQNSVNAGNFSGDD316 pKa = 3.2YY317 pKa = 9.11PATVQGQADD326 pKa = 5.07LIHH329 pKa = 7.17DD330 pKa = 4.06VMAAVAAVGDD340 pKa = 3.81AGLGVFYY347 pKa = 10.3WEE349 pKa = 4.56PAWIPVGTPDD359 pKa = 4.81QLAANQQKK367 pKa = 8.64WEE369 pKa = 4.07QYY371 pKa = 8.49GSGWASSYY379 pKa = 10.97AGSYY383 pKa = 10.89DD384 pKa = 3.57PVDD387 pKa = 3.46AGVYY391 pKa = 9.97YY392 pKa = 10.83GGSSWDD398 pKa = 3.53NQALFDD404 pKa = 4.15FTGHH408 pKa = 6.87PLEE411 pKa = 4.4SLQTFRR417 pKa = 11.84YY418 pKa = 9.22ARR420 pKa = 11.84TGAVTEE426 pKa = 4.38RR427 pKa = 11.84VATSIEE433 pKa = 4.44TVALTVYY440 pKa = 10.22DD441 pKa = 4.43GSPIALPATIEE452 pKa = 3.8VGYY455 pKa = 10.92NDD457 pKa = 4.64GSTEE461 pKa = 3.99HH462 pKa = 7.07PAVAWSDD469 pKa = 3.34AVDD472 pKa = 4.51WIRR475 pKa = 11.84GPGVYY480 pKa = 10.25SIPGTIASGEE490 pKa = 4.36AITATVTVLQANYY503 pKa = 9.66VVNHH507 pKa = 6.2GFEE510 pKa = 4.62DD511 pKa = 4.65ADD513 pKa = 3.5TSMWSLATNPSGAASITTTSNTNEE537 pKa = 3.89GTKK540 pKa = 10.77ALDD543 pKa = 3.66FWAASAFTIQLQQTVTGVPAGEE565 pKa = 4.13YY566 pKa = 9.27TLQATAQGGALGDD579 pKa = 4.05SDD581 pKa = 5.48TLTLWAYY588 pKa = 9.78TDD590 pKa = 4.97DD591 pKa = 5.24DD592 pKa = 5.56SADD595 pKa = 3.1TWMWLYY601 pKa = 10.9GWRR604 pKa = 11.84DD605 pKa = 3.35YY606 pKa = 11.02RR607 pKa = 11.84TFTTPPLTVGDD618 pKa = 4.72DD619 pKa = 3.7GTVTVGAYY627 pKa = 9.63ISATGGAWGTLDD639 pKa = 3.93DD640 pKa = 4.64LRR642 pKa = 11.84LVRR645 pKa = 11.84YY646 pKa = 9.81IPDD649 pKa = 3.62AALDD653 pKa = 3.7TGALEE658 pKa = 4.21EE659 pKa = 4.99LLDD662 pKa = 4.13EE663 pKa = 5.17ADD665 pKa = 3.83GVDD668 pKa = 3.0RR669 pKa = 11.84ALYY672 pKa = 9.93TEE674 pKa = 5.15GSLGGLDD681 pKa = 4.08DD682 pKa = 4.93AVASAQVVLAGSGDD696 pKa = 3.74QDD698 pKa = 4.22DD699 pKa = 4.14VDD701 pKa = 4.22AVVALLRR708 pKa = 11.84AALDD712 pKa = 3.92GLALAPSGEE721 pKa = 4.21PSGEE725 pKa = 3.81PSGEE729 pKa = 3.96PSEE732 pKa = 5.02GPVDD736 pKa = 3.66APVITLGAGTVTPGGTLHH754 pKa = 5.59VTVSGLEE761 pKa = 3.89EE762 pKa = 4.2GSIEE766 pKa = 3.9IGVASTYY773 pKa = 10.36RR774 pKa = 11.84ALATASVVAGTASATVTIPADD795 pKa = 3.65LEE797 pKa = 4.5AGLHH801 pKa = 6.16HH802 pKa = 6.33IQVRR806 pKa = 11.84DD807 pKa = 3.59GSGALLAEE815 pKa = 4.36AEE817 pKa = 4.35LRR819 pKa = 11.84VLALGDD825 pKa = 3.43TGADD829 pKa = 3.19EE830 pKa = 4.24TVIRR834 pKa = 11.84VLAALGAVLVLGGAAAFAWGRR855 pKa = 11.84RR856 pKa = 11.84RR857 pKa = 11.84AAAA860 pKa = 3.8
MM1 pKa = 7.33HH2 pKa = 7.23AHH4 pKa = 7.56SIRR7 pKa = 11.84TGLGIGAVTAVALALVGIPSAIAAADD33 pKa = 4.05GPVEE37 pKa = 4.04AGIFVDD43 pKa = 5.99RR44 pKa = 11.84IDD46 pKa = 4.1GLGDD50 pKa = 3.53DD51 pKa = 6.05FINGVDD57 pKa = 3.55VSSILSEE64 pKa = 4.15EE65 pKa = 4.19EE66 pKa = 4.02SGVVYY71 pKa = 10.86YY72 pKa = 9.45DD73 pKa = 3.91TEE75 pKa = 4.06GHH77 pKa = 5.8QADD80 pKa = 4.62LFDD83 pKa = 3.9VLEE86 pKa = 4.86DD87 pKa = 3.53ADD89 pKa = 3.7INYY92 pKa = 9.17VRR94 pKa = 11.84VRR96 pKa = 11.84VWDD99 pKa = 3.94DD100 pKa = 3.55PYY102 pKa = 11.28DD103 pKa = 4.16AEE105 pKa = 4.58GNGYY109 pKa = 10.66GGGTTDD115 pKa = 4.07LARR118 pKa = 11.84AVEE121 pKa = 3.99IGEE124 pKa = 4.14RR125 pKa = 11.84ATAHH129 pKa = 5.76GMRR132 pKa = 11.84VLVDD136 pKa = 4.63FHH138 pKa = 8.88YY139 pKa = 11.36SDD141 pKa = 4.96FWADD145 pKa = 3.24PAKK148 pKa = 10.54QHH150 pKa = 6.15VPKK153 pKa = 10.36AWEE156 pKa = 5.08GYY158 pKa = 5.39TTEE161 pKa = 4.21QLVTALGAYY170 pKa = 8.85TEE172 pKa = 4.49ASLDD176 pKa = 3.51AFADD180 pKa = 3.59AGVDD184 pKa = 3.26VGMVQIGNEE193 pKa = 4.27TNNTIAGVADD203 pKa = 3.82WDD205 pKa = 3.87GKK207 pKa = 11.07AAMFSAGSAAVRR219 pKa = 11.84EE220 pKa = 4.41VFPDD224 pKa = 3.43ALVALHH230 pKa = 6.12FTNPEE235 pKa = 3.73TPNRR239 pKa = 11.84YY240 pKa = 7.64TGYY243 pKa = 10.17AAQLAAHH250 pKa = 6.74DD251 pKa = 3.65VDD253 pKa = 4.06YY254 pKa = 11.5DD255 pKa = 3.9VFASSYY261 pKa = 8.41YY262 pKa = 10.06NFWHH266 pKa = 7.01GSPEE270 pKa = 3.94NLTSVLSTIADD281 pKa = 3.81SYY283 pKa = 10.9DD284 pKa = 3.37KK285 pKa = 10.99KK286 pKa = 11.66VMVAEE291 pKa = 4.32TSYY294 pKa = 11.72VYY296 pKa = 10.66TFDD299 pKa = 6.26DD300 pKa = 4.01GDD302 pKa = 3.34GWQNSVNAGNFSGDD316 pKa = 3.2YY317 pKa = 9.11PATVQGQADD326 pKa = 5.07LIHH329 pKa = 7.17DD330 pKa = 4.06VMAAVAAVGDD340 pKa = 3.81AGLGVFYY347 pKa = 10.3WEE349 pKa = 4.56PAWIPVGTPDD359 pKa = 4.81QLAANQQKK367 pKa = 8.64WEE369 pKa = 4.07QYY371 pKa = 8.49GSGWASSYY379 pKa = 10.97AGSYY383 pKa = 10.89DD384 pKa = 3.57PVDD387 pKa = 3.46AGVYY391 pKa = 9.97YY392 pKa = 10.83GGSSWDD398 pKa = 3.53NQALFDD404 pKa = 4.15FTGHH408 pKa = 6.87PLEE411 pKa = 4.4SLQTFRR417 pKa = 11.84YY418 pKa = 9.22ARR420 pKa = 11.84TGAVTEE426 pKa = 4.38RR427 pKa = 11.84VATSIEE433 pKa = 4.44TVALTVYY440 pKa = 10.22DD441 pKa = 4.43GSPIALPATIEE452 pKa = 3.8VGYY455 pKa = 10.92NDD457 pKa = 4.64GSTEE461 pKa = 3.99HH462 pKa = 7.07PAVAWSDD469 pKa = 3.34AVDD472 pKa = 4.51WIRR475 pKa = 11.84GPGVYY480 pKa = 10.25SIPGTIASGEE490 pKa = 4.36AITATVTVLQANYY503 pKa = 9.66VVNHH507 pKa = 6.2GFEE510 pKa = 4.62DD511 pKa = 4.65ADD513 pKa = 3.5TSMWSLATNPSGAASITTTSNTNEE537 pKa = 3.89GTKK540 pKa = 10.77ALDD543 pKa = 3.66FWAASAFTIQLQQTVTGVPAGEE565 pKa = 4.13YY566 pKa = 9.27TLQATAQGGALGDD579 pKa = 4.05SDD581 pKa = 5.48TLTLWAYY588 pKa = 9.78TDD590 pKa = 4.97DD591 pKa = 5.24DD592 pKa = 5.56SADD595 pKa = 3.1TWMWLYY601 pKa = 10.9GWRR604 pKa = 11.84DD605 pKa = 3.35YY606 pKa = 11.02RR607 pKa = 11.84TFTTPPLTVGDD618 pKa = 4.72DD619 pKa = 3.7GTVTVGAYY627 pKa = 9.63ISATGGAWGTLDD639 pKa = 3.93DD640 pKa = 4.64LRR642 pKa = 11.84LVRR645 pKa = 11.84YY646 pKa = 9.81IPDD649 pKa = 3.62AALDD653 pKa = 3.7TGALEE658 pKa = 4.21EE659 pKa = 4.99LLDD662 pKa = 4.13EE663 pKa = 5.17ADD665 pKa = 3.83GVDD668 pKa = 3.0RR669 pKa = 11.84ALYY672 pKa = 9.93TEE674 pKa = 5.15GSLGGLDD681 pKa = 4.08DD682 pKa = 4.93AVASAQVVLAGSGDD696 pKa = 3.74QDD698 pKa = 4.22DD699 pKa = 4.14VDD701 pKa = 4.22AVVALLRR708 pKa = 11.84AALDD712 pKa = 3.92GLALAPSGEE721 pKa = 4.21PSGEE725 pKa = 3.81PSGEE729 pKa = 3.96PSEE732 pKa = 5.02GPVDD736 pKa = 3.66APVITLGAGTVTPGGTLHH754 pKa = 5.59VTVSGLEE761 pKa = 3.89EE762 pKa = 4.2GSIEE766 pKa = 3.9IGVASTYY773 pKa = 10.36RR774 pKa = 11.84ALATASVVAGTASATVTIPADD795 pKa = 3.65LEE797 pKa = 4.5AGLHH801 pKa = 6.16HH802 pKa = 6.33IQVRR806 pKa = 11.84DD807 pKa = 3.59GSGALLAEE815 pKa = 4.36AEE817 pKa = 4.35LRR819 pKa = 11.84VLALGDD825 pKa = 3.43TGADD829 pKa = 3.19EE830 pKa = 4.24TVIRR834 pKa = 11.84VLAALGAVLVLGGAAAFAWGRR855 pKa = 11.84RR856 pKa = 11.84RR857 pKa = 11.84AAAA860 pKa = 3.8
Molecular weight: 89.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A387BAS7|A0A387BAS7_9MICO 1 4-dihydroxy-2-naphthoate octaprenyltransferase OS=Protaetiibacter intestinalis OX=2419774 GN=menA PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
943967 |
26 |
9467 |
332.9 |
35.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.027 ± 0.078 | 0.443 ± 0.012 |
5.983 ± 0.035 | 5.888 ± 0.05 |
3.11 ± 0.032 | 9.184 ± 0.054 |
1.9 ± 0.023 | 4.346 ± 0.034 |
1.776 ± 0.035 | 10.514 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.557 ± 0.02 | 1.8 ± 0.024 |
5.488 ± 0.043 | 2.601 ± 0.024 |
7.414 ± 0.072 | 5.288 ± 0.058 |
5.944 ± 0.073 | 9.241 ± 0.045 |
1.5 ± 0.024 | 1.997 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
