
Harryflintia acetispora
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Harryflintia
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
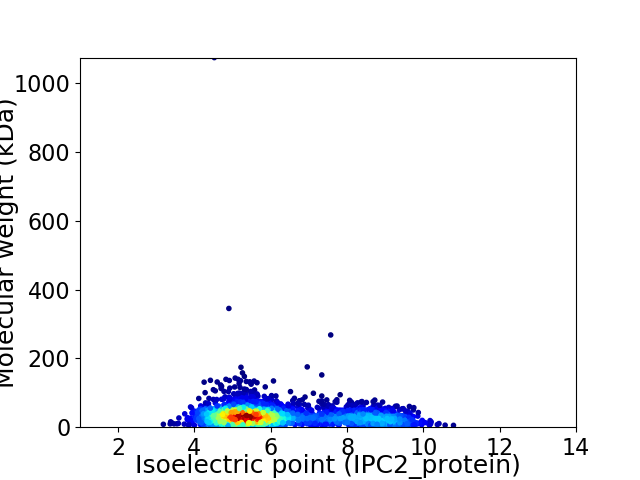
Virtual 2D-PAGE plot for 2808 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3E2STY8|A0A3E2STY8_9FIRM Uncharacterized protein OS=Harryflintia acetispora OX=1849041 GN=DW086_10200 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 10.43KK3 pKa = 10.6VMATLLAGCLLLGSVLSASAASISKK28 pKa = 10.26SSSSVSGYY36 pKa = 10.74SLTVGPGDD44 pKa = 4.77DD45 pKa = 3.48NAEE48 pKa = 4.19YY49 pKa = 10.92SDD51 pKa = 4.34GAPEE55 pKa = 4.32IYY57 pKa = 9.86TYY59 pKa = 10.11GTDD62 pKa = 3.21SMKK65 pKa = 10.41EE66 pKa = 3.79AEE68 pKa = 4.14IAIIPEE74 pKa = 3.96SGYY77 pKa = 10.8EE78 pKa = 3.8ITDD81 pKa = 2.92VDD83 pKa = 4.17VSVNTSNISASYY95 pKa = 9.64KK96 pKa = 10.75KK97 pKa = 10.37NDD99 pKa = 3.59GTVSDD104 pKa = 4.88PGVLTIKK111 pKa = 10.46GKK113 pKa = 10.07AGVRR117 pKa = 11.84VFEE120 pKa = 4.63LEE122 pKa = 4.83DD123 pKa = 3.61YY124 pKa = 11.13EE125 pKa = 5.91LDD127 pKa = 4.44LDD129 pKa = 4.05ITLEE133 pKa = 4.35DD134 pKa = 4.53DD135 pKa = 3.66ATGDD139 pKa = 3.81EE140 pKa = 4.51YY141 pKa = 11.55DD142 pKa = 3.66VTITIYY148 pKa = 11.03GEE150 pKa = 3.83EE151 pKa = 4.05LTAYY155 pKa = 9.74SPSRR159 pKa = 11.84EE160 pKa = 4.16VEE162 pKa = 3.67SDD164 pKa = 2.94EE165 pKa = 4.72TYY167 pKa = 10.59TITKK171 pKa = 10.05SLGCIFEE178 pKa = 3.99FDD180 pKa = 3.69EE181 pKa = 5.35YY182 pKa = 10.54IDD184 pKa = 3.59EE185 pKa = 4.25EE186 pKa = 4.3TRR188 pKa = 11.84IRR190 pKa = 11.84VDD192 pKa = 4.67DD193 pKa = 6.03DD194 pKa = 3.37IDD196 pKa = 4.57LYY198 pKa = 11.74FNGNYY203 pKa = 7.95GTDD206 pKa = 3.65YY207 pKa = 11.22EE208 pKa = 4.29NLRR211 pKa = 11.84VVTDD215 pKa = 4.54SISEE219 pKa = 3.88IDD221 pKa = 3.56KK222 pKa = 11.02ALSDD226 pKa = 4.63LYY228 pKa = 11.28VDD230 pKa = 4.23YY231 pKa = 11.3YY232 pKa = 11.7DD233 pKa = 6.35FIASPKK239 pKa = 8.73FASAVEE245 pKa = 4.04VEE247 pKa = 4.55IKK249 pKa = 10.65AGTEE253 pKa = 3.86AYY255 pKa = 9.51IYY257 pKa = 9.87EE258 pKa = 4.35YY259 pKa = 10.98NKK261 pKa = 9.31KK262 pKa = 8.67TGDD265 pKa = 3.48LTLVDD270 pKa = 5.08AEE272 pKa = 4.7YY273 pKa = 10.94DD274 pKa = 3.88GNWIFEE280 pKa = 4.33TKK282 pKa = 9.82TLGTYY287 pKa = 9.92IVTDD291 pKa = 3.66EE292 pKa = 5.16EE293 pKa = 4.43IDD295 pKa = 3.57EE296 pKa = 4.71SMLSTSTADD305 pKa = 3.68EE306 pKa = 4.99DD307 pKa = 4.1STDD310 pKa = 3.37DD311 pKa = 3.87TEE313 pKa = 5.34YY314 pKa = 11.61DD315 pKa = 3.88DD316 pKa = 4.55VDD318 pKa = 4.04SVSDD322 pKa = 3.97SEE324 pKa = 4.41KK325 pKa = 10.17TNPGTGAAGSPALAVLLGIGSLCAAGLAAYY355 pKa = 9.49RR356 pKa = 11.84KK357 pKa = 9.36AARR360 pKa = 3.98
MM1 pKa = 7.28KK2 pKa = 10.43KK3 pKa = 10.6VMATLLAGCLLLGSVLSASAASISKK28 pKa = 10.26SSSSVSGYY36 pKa = 10.74SLTVGPGDD44 pKa = 4.77DD45 pKa = 3.48NAEE48 pKa = 4.19YY49 pKa = 10.92SDD51 pKa = 4.34GAPEE55 pKa = 4.32IYY57 pKa = 9.86TYY59 pKa = 10.11GTDD62 pKa = 3.21SMKK65 pKa = 10.41EE66 pKa = 3.79AEE68 pKa = 4.14IAIIPEE74 pKa = 3.96SGYY77 pKa = 10.8EE78 pKa = 3.8ITDD81 pKa = 2.92VDD83 pKa = 4.17VSVNTSNISASYY95 pKa = 9.64KK96 pKa = 10.75KK97 pKa = 10.37NDD99 pKa = 3.59GTVSDD104 pKa = 4.88PGVLTIKK111 pKa = 10.46GKK113 pKa = 10.07AGVRR117 pKa = 11.84VFEE120 pKa = 4.63LEE122 pKa = 4.83DD123 pKa = 3.61YY124 pKa = 11.13EE125 pKa = 5.91LDD127 pKa = 4.44LDD129 pKa = 4.05ITLEE133 pKa = 4.35DD134 pKa = 4.53DD135 pKa = 3.66ATGDD139 pKa = 3.81EE140 pKa = 4.51YY141 pKa = 11.55DD142 pKa = 3.66VTITIYY148 pKa = 11.03GEE150 pKa = 3.83EE151 pKa = 4.05LTAYY155 pKa = 9.74SPSRR159 pKa = 11.84EE160 pKa = 4.16VEE162 pKa = 3.67SDD164 pKa = 2.94EE165 pKa = 4.72TYY167 pKa = 10.59TITKK171 pKa = 10.05SLGCIFEE178 pKa = 3.99FDD180 pKa = 3.69EE181 pKa = 5.35YY182 pKa = 10.54IDD184 pKa = 3.59EE185 pKa = 4.25EE186 pKa = 4.3TRR188 pKa = 11.84IRR190 pKa = 11.84VDD192 pKa = 4.67DD193 pKa = 6.03DD194 pKa = 3.37IDD196 pKa = 4.57LYY198 pKa = 11.74FNGNYY203 pKa = 7.95GTDD206 pKa = 3.65YY207 pKa = 11.22EE208 pKa = 4.29NLRR211 pKa = 11.84VVTDD215 pKa = 4.54SISEE219 pKa = 3.88IDD221 pKa = 3.56KK222 pKa = 11.02ALSDD226 pKa = 4.63LYY228 pKa = 11.28VDD230 pKa = 4.23YY231 pKa = 11.3YY232 pKa = 11.7DD233 pKa = 6.35FIASPKK239 pKa = 8.73FASAVEE245 pKa = 4.04VEE247 pKa = 4.55IKK249 pKa = 10.65AGTEE253 pKa = 3.86AYY255 pKa = 9.51IYY257 pKa = 9.87EE258 pKa = 4.35YY259 pKa = 10.98NKK261 pKa = 9.31KK262 pKa = 8.67TGDD265 pKa = 3.48LTLVDD270 pKa = 5.08AEE272 pKa = 4.7YY273 pKa = 10.94DD274 pKa = 3.88GNWIFEE280 pKa = 4.33TKK282 pKa = 9.82TLGTYY287 pKa = 9.92IVTDD291 pKa = 3.66EE292 pKa = 5.16EE293 pKa = 4.43IDD295 pKa = 3.57EE296 pKa = 4.71SMLSTSTADD305 pKa = 3.68EE306 pKa = 4.99DD307 pKa = 4.1STDD310 pKa = 3.37DD311 pKa = 3.87TEE313 pKa = 5.34YY314 pKa = 11.61DD315 pKa = 3.88DD316 pKa = 4.55VDD318 pKa = 4.04SVSDD322 pKa = 3.97SEE324 pKa = 4.41KK325 pKa = 10.17TNPGTGAAGSPALAVLLGIGSLCAAGLAAYY355 pKa = 9.49RR356 pKa = 11.84KK357 pKa = 9.36AARR360 pKa = 3.98
Molecular weight: 38.91 kDa
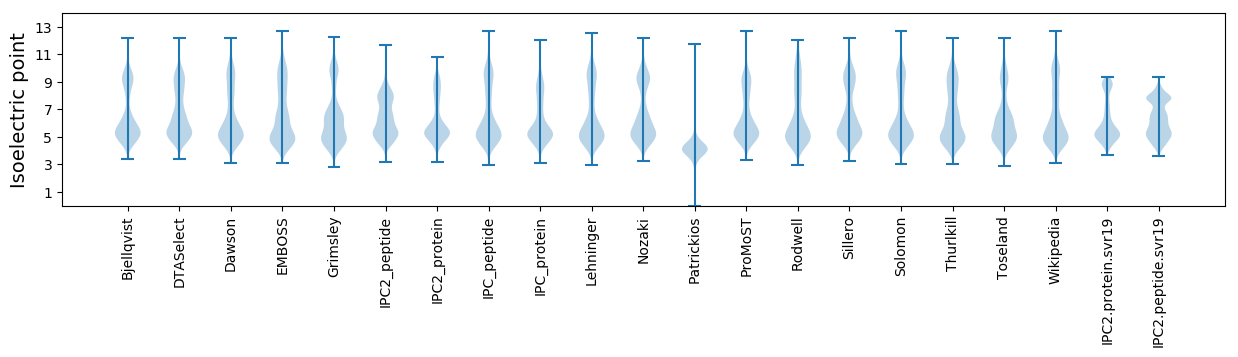
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3E2SYB4|A0A3E2SYB4_9FIRM Type IV secretory system conjugative DNA transfer family protein OS=Harryflintia acetispora OX=1849041 GN=DW086_07850 PE=3 SV=1
MM1 pKa = 7.62GGTLRR6 pKa = 11.84RR7 pKa = 11.84TSKK10 pKa = 7.46TTLWSYY16 pKa = 11.37DD17 pKa = 2.92SRR19 pKa = 11.84ASGDD23 pKa = 3.43RR24 pKa = 11.84NVPIARR30 pKa = 11.84GAWQMWRR37 pKa = 11.84VNFSKK42 pKa = 10.6SDD44 pKa = 3.31TMRR47 pKa = 11.84WQLRR51 pKa = 11.84LPAHH55 pKa = 5.91SCDD58 pKa = 3.76FEE60 pKa = 5.4SNNHH64 pKa = 5.17IFDD67 pKa = 4.04RR68 pKa = 11.84KK69 pKa = 10.34GDD71 pKa = 4.11HH72 pKa = 6.04LWKK75 pKa = 10.82NN76 pKa = 3.6
MM1 pKa = 7.62GGTLRR6 pKa = 11.84RR7 pKa = 11.84TSKK10 pKa = 7.46TTLWSYY16 pKa = 11.37DD17 pKa = 2.92SRR19 pKa = 11.84ASGDD23 pKa = 3.43RR24 pKa = 11.84NVPIARR30 pKa = 11.84GAWQMWRR37 pKa = 11.84VNFSKK42 pKa = 10.6SDD44 pKa = 3.31TMRR47 pKa = 11.84WQLRR51 pKa = 11.84LPAHH55 pKa = 5.91SCDD58 pKa = 3.76FEE60 pKa = 5.4SNNHH64 pKa = 5.17IFDD67 pKa = 4.04RR68 pKa = 11.84KK69 pKa = 10.34GDD71 pKa = 4.11HH72 pKa = 6.04LWKK75 pKa = 10.82NN76 pKa = 3.6
Molecular weight: 8.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
875103 |
27 |
10091 |
311.6 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.157 ± 0.043 | 1.736 ± 0.026 |
5.23 ± 0.038 | 6.975 ± 0.057 |
3.945 ± 0.038 | 8.336 ± 0.046 |
1.727 ± 0.02 | 5.85 ± 0.046 |
4.763 ± 0.046 | 10.23 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.672 ± 0.03 | 3.171 ± 0.033 |
4.241 ± 0.03 | 3.481 ± 0.03 |
5.852 ± 0.052 | 5.997 ± 0.049 |
5.055 ± 0.056 | 7.052 ± 0.041 |
0.921 ± 0.015 | 3.609 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
