
Streptomyces sp. MZ03-48
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
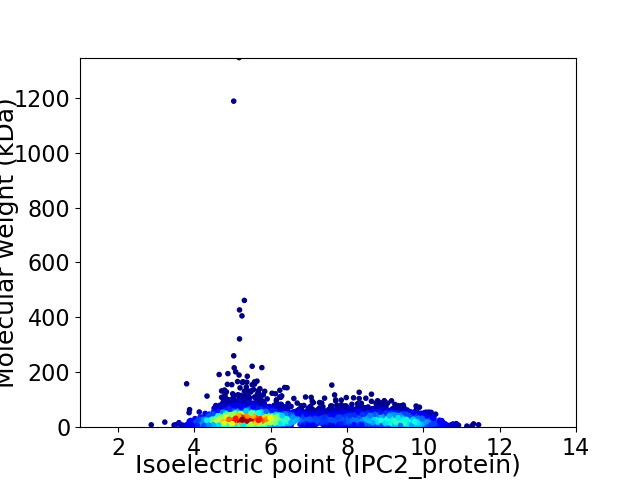
Virtual 2D-PAGE plot for 5822 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553ZQS1|A0A553ZQS1_9ACTN Sec-independent protein translocase protein TatA OS=Streptomyces sp. MZ03-48 OX=2593677 GN=tatA PE=3 SV=1
MM1 pKa = 7.66GLRR4 pKa = 11.84TPAIPLSAGRR14 pKa = 11.84GCTSVQPASRR24 pKa = 11.84QPIQIRR30 pKa = 11.84QTGDD34 pKa = 3.01LEE36 pKa = 4.11GRR38 pKa = 11.84IMAKK42 pKa = 10.17AFQPSWRR49 pKa = 11.84QLVGLPWGGSHH60 pKa = 6.98RR61 pKa = 11.84PLAVVSSDD69 pKa = 3.07PTVTVGTTPTGVAITPDD86 pKa = 3.35GLHH89 pKa = 6.86AYY91 pKa = 8.44VANEE95 pKa = 3.65GSNDD99 pKa = 3.17VSVIDD104 pKa = 3.92TTSLTVTATVPVGTTPFWVAITPDD128 pKa = 3.36GLRR131 pKa = 11.84AYY133 pKa = 7.79VTNASDD139 pKa = 3.7GTVSVIDD146 pKa = 3.76TVTNTVTATIPVGTTPLGVAIGGTSAYY173 pKa = 8.02VTNASDD179 pKa = 3.7GTVSVIDD186 pKa = 3.76TVTNTVTATIPVGTAPFGVAIGGTSAYY213 pKa = 8.15VTNNGDD219 pKa = 3.48GTVSVIDD226 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH251 pKa = 7.05AYY253 pKa = 8.99VANNGDD259 pKa = 3.68GTVSVIDD266 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH291 pKa = 7.05AYY293 pKa = 8.99VANNGDD299 pKa = 3.68GTVSVIDD306 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH331 pKa = 7.05AYY333 pKa = 8.99VANNGDD339 pKa = 3.68GTVSVIDD346 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH371 pKa = 7.05AYY373 pKa = 8.99VANNGDD379 pKa = 3.79DD380 pKa = 3.78TVSVIDD386 pKa = 3.71TGTNTVTFTIPVGPGPFGVAITPDD410 pKa = 3.03GSTIYY415 pKa = 10.88VADD418 pKa = 3.77NADD421 pKa = 3.39NSVTVTAAVPFPTTTVLTSAPDD443 pKa = 3.58PSVFGEE449 pKa = 4.2AKK451 pKa = 9.75TLTATVTSSSGTPTGTVGFFDD472 pKa = 4.43GATLLGTGTLSGGVATFTTSALAVGSHH499 pKa = 6.87ALTAVYY505 pKa = 10.34GGDD508 pKa = 3.46TNFAGSTSPVDD519 pKa = 3.62TQTVGQAVTSTALSSAPDD537 pKa = 3.57PSVFGEE543 pKa = 4.2AKK545 pKa = 9.75TLTATVTVVAPGAGTPTGTVSFFDD569 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH596 pKa = 6.78ALTAVYY602 pKa = 10.57SGDD605 pKa = 3.35TDD607 pKa = 4.03FAGSTSPVDD616 pKa = 3.62TQTVGQAVTTTALSSAPDD634 pKa = 3.57PSVFGEE640 pKa = 4.2AKK642 pKa = 9.75TLTATVTVVAPGAGTLTGTVSFFDD666 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH693 pKa = 6.78ALTAVYY699 pKa = 10.47SGDD702 pKa = 3.85SNFTGSTSPVDD713 pKa = 3.63TQTVNQAATTTALTSAPDD731 pKa = 3.62PSVFGEE737 pKa = 4.2AKK739 pKa = 9.75TLTATVTVVAPGAGTPTGTVSFFDD763 pKa = 3.97GATLLGTGTLSGGVASFTTSALAVGSHH790 pKa = 6.87ALTAVYY796 pKa = 10.34GGDD799 pKa = 3.46TNFAGSTSPVDD810 pKa = 3.62TQTVGQAVTTTALSSAPDD828 pKa = 3.57PSVFGEE834 pKa = 4.2AKK836 pKa = 9.75TLTATVTVLAPGVGTPTGTVSFFDD860 pKa = 3.97GATLLGTGTLSGGVASFTTSTLGIGSHH887 pKa = 6.79ALTAVYY893 pKa = 10.43SGDD896 pKa = 3.64TNFVGSTSPVDD907 pKa = 3.63TQTVGQAATTTALSSAPDD925 pKa = 3.57PSVFGEE931 pKa = 4.2AKK933 pKa = 9.66TLTATVSAVSPGSGTPTGTVSFFDD957 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH984 pKa = 6.78ALTAVYY990 pKa = 10.57SGDD993 pKa = 3.35TDD995 pKa = 4.03FAGSTSPVDD1004 pKa = 3.58TQTVNQAATTTVLSSAPDD1022 pKa = 3.54PSVFGEE1028 pKa = 4.2AKK1030 pKa = 9.75TLTATVTVVAPGAGTLTGTVSFFDD1054 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH1081 pKa = 6.78ALTAVYY1087 pKa = 10.47SGDD1090 pKa = 3.85SNFTGSTSPVDD1101 pKa = 3.68TQTVGQAATSTALSSAPDD1119 pKa = 3.57PSVFGEE1125 pKa = 4.2AKK1127 pKa = 9.75TLTATVTVVAPGVGTPTGTVGFFDD1151 pKa = 4.43GATLLGTGTLSGGVASFTTSSLSVGSHH1178 pKa = 6.78ALTAVYY1184 pKa = 10.58SGDD1187 pKa = 3.41TDD1189 pKa = 3.42IAGSTSPVDD1198 pKa = 3.62TQTVGQAATSTALSSAPDD1216 pKa = 3.57PSVFGEE1222 pKa = 4.2AKK1224 pKa = 9.75TLTATVTVVAPGVGTPTGTVGFFDD1248 pKa = 4.43GATLLGTGTLSGGVATFTTSTLGIGSHH1275 pKa = 6.79ALTAVYY1281 pKa = 10.43SGDD1284 pKa = 3.64TNFVGSTSPVDD1295 pKa = 3.61TQTVNQAATSTALSSAPDD1313 pKa = 3.57PSVFGEE1319 pKa = 4.2AKK1321 pKa = 9.75TLTATVTVVAPGVGTPTGTVSFFDD1345 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH1372 pKa = 6.78ALTAVYY1378 pKa = 10.36SGDD1381 pKa = 3.15IDD1383 pKa = 4.21FVGSTSPVDD1392 pKa = 3.63TQTVGQAATTTALTSAPDD1410 pKa = 3.69PSVFGQAKK1418 pKa = 8.14TLTATVSAVAPGAGTPTGTVSFFDD1442 pKa = 3.97GATLLGTGTLSGGVATFTTSSLSVGSHH1469 pKa = 6.78ALTAVYY1475 pKa = 10.47SGDD1478 pKa = 3.85SNFTGSTSPVDD1489 pKa = 3.63TQTVNQAATTTALSSAPDD1507 pKa = 3.68PSVFGQAKK1515 pKa = 8.22TLTATVTAVPPGSGTPTGTVSFFDD1539 pKa = 3.97GATLLGTGTLSGGVASFTTSTLGIGTHH1566 pKa = 5.96SLTAVYY1572 pKa = 10.32NGSGNFNTSTSPVDD1586 pKa = 3.51TQTVTKK1592 pKa = 10.66NLTTTALTSAPDD1604 pKa = 3.69PSVFGQAKK1612 pKa = 8.22TLTATVTAVPPGSGTPTGTVSFFDD1636 pKa = 3.99GATLLGG1642 pKa = 3.94
MM1 pKa = 7.66GLRR4 pKa = 11.84TPAIPLSAGRR14 pKa = 11.84GCTSVQPASRR24 pKa = 11.84QPIQIRR30 pKa = 11.84QTGDD34 pKa = 3.01LEE36 pKa = 4.11GRR38 pKa = 11.84IMAKK42 pKa = 10.17AFQPSWRR49 pKa = 11.84QLVGLPWGGSHH60 pKa = 6.98RR61 pKa = 11.84PLAVVSSDD69 pKa = 3.07PTVTVGTTPTGVAITPDD86 pKa = 3.35GLHH89 pKa = 6.86AYY91 pKa = 8.44VANEE95 pKa = 3.65GSNDD99 pKa = 3.17VSVIDD104 pKa = 3.92TTSLTVTATVPVGTTPFWVAITPDD128 pKa = 3.36GLRR131 pKa = 11.84AYY133 pKa = 7.79VTNASDD139 pKa = 3.7GTVSVIDD146 pKa = 3.76TVTNTVTATIPVGTTPLGVAIGGTSAYY173 pKa = 8.02VTNASDD179 pKa = 3.7GTVSVIDD186 pKa = 3.76TVTNTVTATIPVGTAPFGVAIGGTSAYY213 pKa = 8.15VTNNGDD219 pKa = 3.48GTVSVIDD226 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH251 pKa = 7.05AYY253 pKa = 8.99VANNGDD259 pKa = 3.68GTVSVIDD266 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH291 pKa = 7.05AYY293 pKa = 8.99VANNGDD299 pKa = 3.68GTVSVIDD306 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH331 pKa = 7.05AYY333 pKa = 8.99VANNGDD339 pKa = 3.68GTVSVIDD346 pKa = 3.6TGTNTVTATVTVGTAPLGVAVAGTHH371 pKa = 7.05AYY373 pKa = 8.99VANNGDD379 pKa = 3.79DD380 pKa = 3.78TVSVIDD386 pKa = 3.71TGTNTVTFTIPVGPGPFGVAITPDD410 pKa = 3.03GSTIYY415 pKa = 10.88VADD418 pKa = 3.77NADD421 pKa = 3.39NSVTVTAAVPFPTTTVLTSAPDD443 pKa = 3.58PSVFGEE449 pKa = 4.2AKK451 pKa = 9.75TLTATVTSSSGTPTGTVGFFDD472 pKa = 4.43GATLLGTGTLSGGVATFTTSALAVGSHH499 pKa = 6.87ALTAVYY505 pKa = 10.34GGDD508 pKa = 3.46TNFAGSTSPVDD519 pKa = 3.62TQTVGQAVTSTALSSAPDD537 pKa = 3.57PSVFGEE543 pKa = 4.2AKK545 pKa = 9.75TLTATVTVVAPGAGTPTGTVSFFDD569 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH596 pKa = 6.78ALTAVYY602 pKa = 10.57SGDD605 pKa = 3.35TDD607 pKa = 4.03FAGSTSPVDD616 pKa = 3.62TQTVGQAVTTTALSSAPDD634 pKa = 3.57PSVFGEE640 pKa = 4.2AKK642 pKa = 9.75TLTATVTVVAPGAGTLTGTVSFFDD666 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH693 pKa = 6.78ALTAVYY699 pKa = 10.47SGDD702 pKa = 3.85SNFTGSTSPVDD713 pKa = 3.63TQTVNQAATTTALTSAPDD731 pKa = 3.62PSVFGEE737 pKa = 4.2AKK739 pKa = 9.75TLTATVTVVAPGAGTPTGTVSFFDD763 pKa = 3.97GATLLGTGTLSGGVASFTTSALAVGSHH790 pKa = 6.87ALTAVYY796 pKa = 10.34GGDD799 pKa = 3.46TNFAGSTSPVDD810 pKa = 3.62TQTVGQAVTTTALSSAPDD828 pKa = 3.57PSVFGEE834 pKa = 4.2AKK836 pKa = 9.75TLTATVTVLAPGVGTPTGTVSFFDD860 pKa = 3.97GATLLGTGTLSGGVASFTTSTLGIGSHH887 pKa = 6.79ALTAVYY893 pKa = 10.43SGDD896 pKa = 3.64TNFVGSTSPVDD907 pKa = 3.63TQTVGQAATTTALSSAPDD925 pKa = 3.57PSVFGEE931 pKa = 4.2AKK933 pKa = 9.66TLTATVSAVSPGSGTPTGTVSFFDD957 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH984 pKa = 6.78ALTAVYY990 pKa = 10.57SGDD993 pKa = 3.35TDD995 pKa = 4.03FAGSTSPVDD1004 pKa = 3.58TQTVNQAATTTVLSSAPDD1022 pKa = 3.54PSVFGEE1028 pKa = 4.2AKK1030 pKa = 9.75TLTATVTVVAPGAGTLTGTVSFFDD1054 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH1081 pKa = 6.78ALTAVYY1087 pKa = 10.47SGDD1090 pKa = 3.85SNFTGSTSPVDD1101 pKa = 3.68TQTVGQAATSTALSSAPDD1119 pKa = 3.57PSVFGEE1125 pKa = 4.2AKK1127 pKa = 9.75TLTATVTVVAPGVGTPTGTVGFFDD1151 pKa = 4.43GATLLGTGTLSGGVASFTTSSLSVGSHH1178 pKa = 6.78ALTAVYY1184 pKa = 10.58SGDD1187 pKa = 3.41TDD1189 pKa = 3.42IAGSTSPVDD1198 pKa = 3.62TQTVGQAATSTALSSAPDD1216 pKa = 3.57PSVFGEE1222 pKa = 4.2AKK1224 pKa = 9.75TLTATVTVVAPGVGTPTGTVGFFDD1248 pKa = 4.43GATLLGTGTLSGGVATFTTSTLGIGSHH1275 pKa = 6.79ALTAVYY1281 pKa = 10.43SGDD1284 pKa = 3.64TNFVGSTSPVDD1295 pKa = 3.61TQTVNQAATSTALSSAPDD1313 pKa = 3.57PSVFGEE1319 pKa = 4.2AKK1321 pKa = 9.75TLTATVTVVAPGVGTPTGTVSFFDD1345 pKa = 3.97GATLLGTGTLSGGVASFTTSSLSVGSHH1372 pKa = 6.78ALTAVYY1378 pKa = 10.36SGDD1381 pKa = 3.15IDD1383 pKa = 4.21FVGSTSPVDD1392 pKa = 3.63TQTVGQAATTTALTSAPDD1410 pKa = 3.69PSVFGQAKK1418 pKa = 8.14TLTATVSAVAPGAGTPTGTVSFFDD1442 pKa = 3.97GATLLGTGTLSGGVATFTTSSLSVGSHH1469 pKa = 6.78ALTAVYY1475 pKa = 10.47SGDD1478 pKa = 3.85SNFTGSTSPVDD1489 pKa = 3.63TQTVNQAATTTALSSAPDD1507 pKa = 3.68PSVFGQAKK1515 pKa = 8.22TLTATVTAVPPGSGTPTGTVSFFDD1539 pKa = 3.97GATLLGTGTLSGGVASFTTSTLGIGTHH1566 pKa = 5.96SLTAVYY1572 pKa = 10.32NGSGNFNTSTSPVDD1586 pKa = 3.51TQTVTKK1592 pKa = 10.66NLTTTALTSAPDD1604 pKa = 3.69PSVFGQAKK1612 pKa = 8.22TLTATVTAVPPGSGTPTGTVSFFDD1636 pKa = 3.99GATLLGG1642 pKa = 3.94
Molecular weight: 157.99 kDa
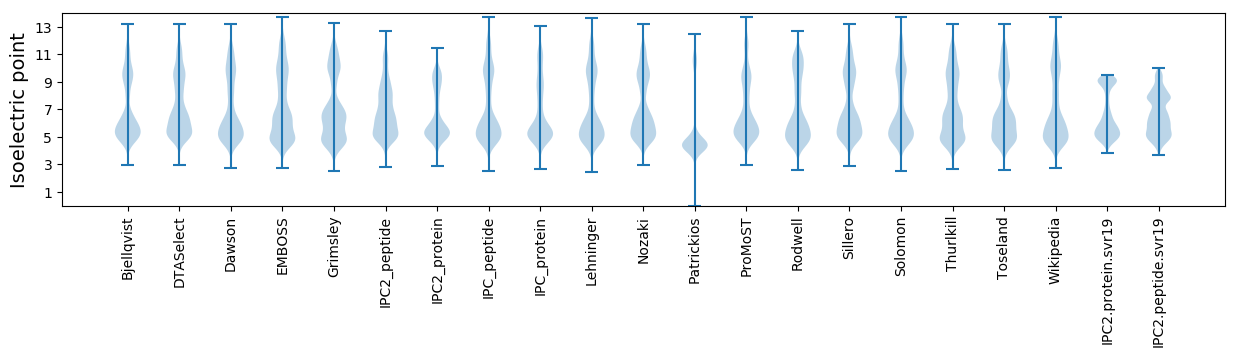
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553YVP9|A0A553YVP9_9ACTN Maltose alpha-D-glucosyltransferase OS=Streptomyces sp. MZ03-48 OX=2593677 GN=treS PE=3 SV=1
MM1 pKa = 7.25GHH3 pKa = 7.23LLLRR7 pKa = 11.84GLLRR11 pKa = 11.84TLLLRR16 pKa = 11.84LRR18 pKa = 11.84HH19 pKa = 5.61LRR21 pKa = 11.84GLLRR25 pKa = 11.84LLGLGLLLRR34 pKa = 11.84GRR36 pKa = 11.84RR37 pKa = 11.84LLLRR41 pKa = 11.84RR42 pKa = 11.84LLLRR46 pKa = 11.84LLGVRR51 pKa = 11.84VLLLNLRR58 pKa = 11.84GRR60 pKa = 11.84LLLVRR65 pKa = 11.84AGLLLRR71 pKa = 11.84LRR73 pKa = 11.84RR74 pKa = 11.84LRR76 pKa = 11.84VRR78 pKa = 11.84ARR80 pKa = 11.84LLGLLDD86 pKa = 3.42VRR88 pKa = 11.84VLLGRR93 pKa = 11.84GLLGMRR99 pKa = 11.84ARR101 pKa = 11.84LLRR104 pKa = 4.4
MM1 pKa = 7.25GHH3 pKa = 7.23LLLRR7 pKa = 11.84GLLRR11 pKa = 11.84TLLLRR16 pKa = 11.84LRR18 pKa = 11.84HH19 pKa = 5.61LRR21 pKa = 11.84GLLRR25 pKa = 11.84LLGLGLLLRR34 pKa = 11.84GRR36 pKa = 11.84RR37 pKa = 11.84LLLRR41 pKa = 11.84RR42 pKa = 11.84LLLRR46 pKa = 11.84LLGVRR51 pKa = 11.84VLLLNLRR58 pKa = 11.84GRR60 pKa = 11.84LLLVRR65 pKa = 11.84AGLLLRR71 pKa = 11.84LRR73 pKa = 11.84RR74 pKa = 11.84LRR76 pKa = 11.84VRR78 pKa = 11.84ARR80 pKa = 11.84LLGLLDD86 pKa = 3.42VRR88 pKa = 11.84VLLGRR93 pKa = 11.84GLLGMRR99 pKa = 11.84ARR101 pKa = 11.84LLRR104 pKa = 4.4
Molecular weight: 12.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1838379 |
24 |
12400 |
315.8 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.046 ± 0.054 | 0.827 ± 0.012 |
5.959 ± 0.028 | 5.589 ± 0.037 |
2.675 ± 0.019 | 9.691 ± 0.037 |
2.441 ± 0.018 | 3.07 ± 0.023 |
2.199 ± 0.029 | 10.335 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.718 ± 0.011 | 1.68 ± 0.018 |
6.288 ± 0.032 | 2.718 ± 0.034 |
8.241 ± 0.039 | 4.765 ± 0.028 |
6.168 ± 0.029 | 8.202 ± 0.036 |
1.419 ± 0.014 | 1.97 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
