
Macrococcus sp. DPC7161
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Macrococcus; unclassified Macrococcus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
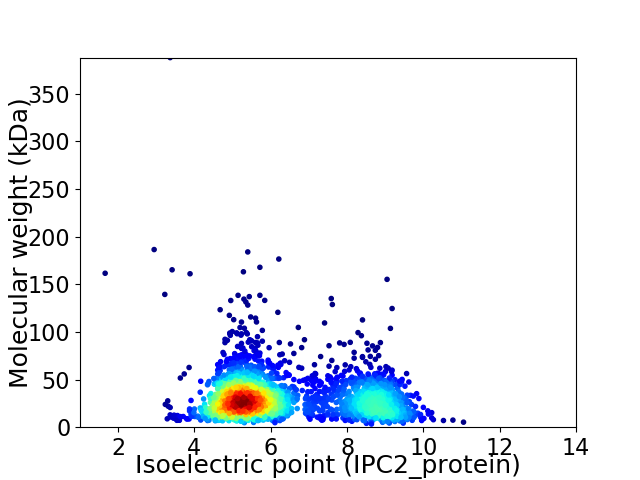
Virtual 2D-PAGE plot for 2341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
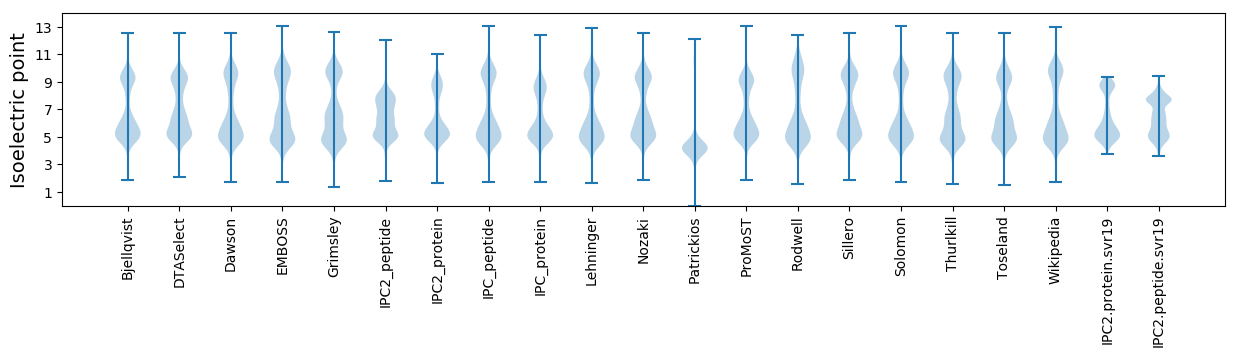
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1M1R4|A0A4V1M1R4_9STAP RNA-binding protein S1 OS=Macrococcus sp. DPC7161 OX=2507060 GN=ER639_11605 PE=4 SV=1
LL1 pKa = 7.59NYY3 pKa = 10.41ASSITSAVEE12 pKa = 3.82RR13 pKa = 11.84VDD15 pKa = 4.39GNPNYY20 pKa = 10.15PIDD23 pKa = 3.95YY24 pKa = 8.57TQVFYY29 pKa = 10.71INPLNKK35 pKa = 9.65TMPATSLEE43 pKa = 4.22VVPYY47 pKa = 10.08HH48 pKa = 6.52PSYY51 pKa = 10.83PNSLAQLDD59 pKa = 3.91PSVTNFHH66 pKa = 6.73IYY68 pKa = 10.42EE69 pKa = 4.18YY70 pKa = 10.77TDD72 pKa = 3.45GSLLNDD78 pKa = 4.04SFSNDD83 pKa = 2.89YY84 pKa = 11.2SLAIDD89 pKa = 3.57VTSQYY94 pKa = 11.34SPIWNEE100 pKa = 3.86DD101 pKa = 3.05GTVTFNFGDD110 pKa = 3.31ITKK113 pKa = 8.85TYY115 pKa = 9.73VVVVDD120 pKa = 4.47SVTKK124 pKa = 10.32VDD126 pKa = 4.04PSNKK130 pKa = 9.26ILVQSGLVSADD141 pKa = 3.49TTQGTHH147 pKa = 4.63VTRR150 pKa = 11.84STANLTQAFSGVSIGSPTFKK170 pKa = 10.49LGNFVWNDD178 pKa = 3.22LNKK181 pKa = 10.76NGIQEE186 pKa = 3.86IGEE189 pKa = 4.07NGVANVTVTITNSNGTVVGTTQTDD213 pKa = 3.62VNGGYY218 pKa = 10.57LFQNLPEE225 pKa = 4.15GTYY228 pKa = 9.17TVEE231 pKa = 4.61FSNIPTGFVVSPSNSGTDD249 pKa = 3.32PSLDD253 pKa = 3.76SNGLISTVTIANGDD267 pKa = 3.89NLTADD272 pKa = 4.08LGIYY276 pKa = 9.38EE277 pKa = 4.98PIYY280 pKa = 10.56RR281 pKa = 11.84IGDD284 pKa = 4.05YY285 pKa = 10.88VWLDD289 pKa = 3.31TDD291 pKa = 3.77GDD293 pKa = 4.67GIQGTSVDD301 pKa = 3.72EE302 pKa = 4.61APFEE306 pKa = 4.26GAQVILYY313 pKa = 9.91DD314 pKa = 3.83EE315 pKa = 4.94NGNKK319 pKa = 9.33IAEE322 pKa = 4.38TLTDD326 pKa = 3.05NRR328 pKa = 11.84GYY330 pKa = 11.44YY331 pKa = 9.78EE332 pKa = 5.51FNNLDD337 pKa = 3.29NGTYY341 pKa = 8.86QVEE344 pKa = 5.28FINPNPDD351 pKa = 2.59MLAFTKK357 pKa = 10.65QNAGADD363 pKa = 3.81NVDD366 pKa = 3.84SDD368 pKa = 5.08VSDD371 pKa = 3.55ANNRR375 pKa = 11.84VTVTINNADD384 pKa = 3.55NMTIDD389 pKa = 4.34AGLQQVYY396 pKa = 10.8SIGDD400 pKa = 3.98KK401 pKa = 10.8VWLDD405 pKa = 3.57TNGDD409 pKa = 4.07DD410 pKa = 3.78IQNGGTEE417 pKa = 3.95PGIAGVQVQLVDD429 pKa = 3.45KK430 pKa = 8.13TTGTVIQTQTTDD442 pKa = 2.99VNGDD446 pKa = 3.61YY447 pKa = 10.87LFTGLKK453 pKa = 9.59PGDD456 pKa = 3.65YY457 pKa = 8.97TVVFGTPAQVTNPDD471 pKa = 3.47GTTTNYY477 pKa = 10.51TEE479 pKa = 4.08VV480 pKa = 3.25
LL1 pKa = 7.59NYY3 pKa = 10.41ASSITSAVEE12 pKa = 3.82RR13 pKa = 11.84VDD15 pKa = 4.39GNPNYY20 pKa = 10.15PIDD23 pKa = 3.95YY24 pKa = 8.57TQVFYY29 pKa = 10.71INPLNKK35 pKa = 9.65TMPATSLEE43 pKa = 4.22VVPYY47 pKa = 10.08HH48 pKa = 6.52PSYY51 pKa = 10.83PNSLAQLDD59 pKa = 3.91PSVTNFHH66 pKa = 6.73IYY68 pKa = 10.42EE69 pKa = 4.18YY70 pKa = 10.77TDD72 pKa = 3.45GSLLNDD78 pKa = 4.04SFSNDD83 pKa = 2.89YY84 pKa = 11.2SLAIDD89 pKa = 3.57VTSQYY94 pKa = 11.34SPIWNEE100 pKa = 3.86DD101 pKa = 3.05GTVTFNFGDD110 pKa = 3.31ITKK113 pKa = 8.85TYY115 pKa = 9.73VVVVDD120 pKa = 4.47SVTKK124 pKa = 10.32VDD126 pKa = 4.04PSNKK130 pKa = 9.26ILVQSGLVSADD141 pKa = 3.49TTQGTHH147 pKa = 4.63VTRR150 pKa = 11.84STANLTQAFSGVSIGSPTFKK170 pKa = 10.49LGNFVWNDD178 pKa = 3.22LNKK181 pKa = 10.76NGIQEE186 pKa = 3.86IGEE189 pKa = 4.07NGVANVTVTITNSNGTVVGTTQTDD213 pKa = 3.62VNGGYY218 pKa = 10.57LFQNLPEE225 pKa = 4.15GTYY228 pKa = 9.17TVEE231 pKa = 4.61FSNIPTGFVVSPSNSGTDD249 pKa = 3.32PSLDD253 pKa = 3.76SNGLISTVTIANGDD267 pKa = 3.89NLTADD272 pKa = 4.08LGIYY276 pKa = 9.38EE277 pKa = 4.98PIYY280 pKa = 10.56RR281 pKa = 11.84IGDD284 pKa = 4.05YY285 pKa = 10.88VWLDD289 pKa = 3.31TDD291 pKa = 3.77GDD293 pKa = 4.67GIQGTSVDD301 pKa = 3.72EE302 pKa = 4.61APFEE306 pKa = 4.26GAQVILYY313 pKa = 9.91DD314 pKa = 3.83EE315 pKa = 4.94NGNKK319 pKa = 9.33IAEE322 pKa = 4.38TLTDD326 pKa = 3.05NRR328 pKa = 11.84GYY330 pKa = 11.44YY331 pKa = 9.78EE332 pKa = 5.51FNNLDD337 pKa = 3.29NGTYY341 pKa = 8.86QVEE344 pKa = 5.28FINPNPDD351 pKa = 2.59MLAFTKK357 pKa = 10.65QNAGADD363 pKa = 3.81NVDD366 pKa = 3.84SDD368 pKa = 5.08VSDD371 pKa = 3.55ANNRR375 pKa = 11.84VTVTINNADD384 pKa = 3.55NMTIDD389 pKa = 4.34AGLQQVYY396 pKa = 10.8SIGDD400 pKa = 3.98KK401 pKa = 10.8VWLDD405 pKa = 3.57TNGDD409 pKa = 4.07DD410 pKa = 3.78IQNGGTEE417 pKa = 3.95PGIAGVQVQLVDD429 pKa = 3.45KK430 pKa = 8.13TTGTVIQTQTTDD442 pKa = 2.99VNGDD446 pKa = 3.61YY447 pKa = 10.87LFTGLKK453 pKa = 9.59PGDD456 pKa = 3.65YY457 pKa = 8.97TVVFGTPAQVTNPDD471 pKa = 3.47GTTTNYY477 pKa = 10.51TEE479 pKa = 4.08VV480 pKa = 3.25
Molecular weight: 51.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q1B3R0|A0A4Q1B3R0_9STAP Uncharacterized protein OS=Macrococcus sp. DPC7161 OX=2507060 GN=ER639_05715 PE=4 SV=1
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.58VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.94GRR40 pKa = 11.84KK41 pKa = 8.67VLSAA45 pKa = 4.05
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.58VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.94GRR40 pKa = 11.84KK41 pKa = 8.67VLSAA45 pKa = 4.05
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
690768 |
27 |
3845 |
295.1 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.154 ± 0.057 | 0.699 ± 0.015 |
5.652 ± 0.069 | 6.83 ± 0.07 |
4.6 ± 0.047 | 6.153 ± 0.056 |
2.383 ± 0.034 | 8.633 ± 0.065 |
7.568 ± 0.065 | 9.336 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.968 ± 0.029 | 5.205 ± 0.045 |
3.289 ± 0.053 | 3.841 ± 0.041 |
3.314 ± 0.045 | 5.907 ± 0.098 |
5.85 ± 0.123 | 6.928 ± 0.069 |
0.725 ± 0.017 | 3.964 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
