
Bacillus wakoensis JCM 9140
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalihalobacillus; Alkalihalobacillus wakoensis
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
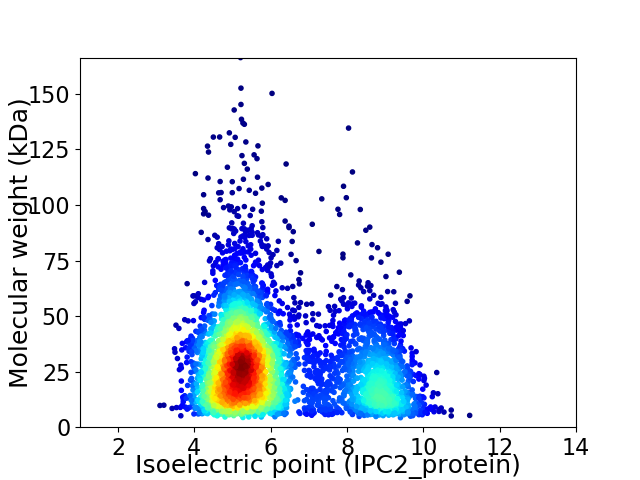
Virtual 2D-PAGE plot for 4575 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4Q8D0|W4Q8D0_9BACI Uncharacterized protein OS=Bacillus wakoensis JCM 9140 OX=1236970 GN=JCM9140_4543 PE=4 SV=1
MM1 pKa = 7.56LKK3 pKa = 10.22KK4 pKa = 10.37KK5 pKa = 9.51WGTYY9 pKa = 8.36VLAGAFTLMMTACGSSTEE27 pKa = 4.29TEE29 pKa = 4.3STSEE33 pKa = 4.24SVDD36 pKa = 3.49APSDD40 pKa = 3.46DD41 pKa = 4.07PEE43 pKa = 4.54PVEE46 pKa = 4.38EE47 pKa = 4.22TVEE50 pKa = 4.09MTDD53 pKa = 3.47VKK55 pKa = 11.03QVTNWFAQPEE65 pKa = 4.39HH66 pKa = 6.56GGLYY70 pKa = 10.45AALEE74 pKa = 4.02KK75 pKa = 10.96GFYY78 pKa = 9.84EE79 pKa = 4.14EE80 pKa = 5.17NGMNMSIDD88 pKa = 3.22QGGPQVSSIQIVASGQAQFGLAQADD113 pKa = 4.29DD114 pKa = 3.7ILIARR119 pKa = 11.84AEE121 pKa = 4.18GVPIVAIGTIFQTSPQGLLFHH142 pKa = 6.96NSQGITDD149 pKa = 4.34FADD152 pKa = 3.35LQGRR156 pKa = 11.84KK157 pKa = 9.96GIIQPGIGYY166 pKa = 8.88WEE168 pKa = 4.38YY169 pKa = 10.71IKK171 pKa = 10.68EE172 pKa = 4.25FYY174 pKa = 11.01DD175 pKa = 4.03LDD177 pKa = 4.1DD178 pKa = 3.8VQEE181 pKa = 4.13ITFTGQFIDD190 pKa = 4.94FIEE193 pKa = 5.41DD194 pKa = 3.29EE195 pKa = 4.41TSFTQGYY202 pKa = 6.57VTQDD206 pKa = 3.22PFNFEE211 pKa = 4.01QQGLDD216 pKa = 3.11TDD218 pKa = 3.86YY219 pKa = 11.91LLIHH223 pKa = 7.34DD224 pKa = 5.09SGYY227 pKa = 10.28QPYY230 pKa = 10.12ANVMFTTEE238 pKa = 4.38EE239 pKa = 4.34LIEE242 pKa = 4.3NNPEE246 pKa = 3.77LVSAYY251 pKa = 10.54VEE253 pKa = 4.09ASVKK257 pKa = 9.89GWEE260 pKa = 4.55FYY262 pKa = 10.71KK263 pKa = 10.83EE264 pKa = 4.18NYY266 pKa = 10.23DD267 pKa = 3.85EE268 pKa = 4.69INPVIQEE275 pKa = 4.28SNPDD279 pKa = 3.42FPTDD283 pKa = 3.41QLAFTAEE290 pKa = 4.2TLVTYY295 pKa = 10.7AFEE298 pKa = 5.17GDD300 pKa = 3.52AAEE303 pKa = 4.68HH304 pKa = 6.19GFGYY308 pKa = 9.93MSEE311 pKa = 4.3EE312 pKa = 3.89RR313 pKa = 11.84WTTLHH318 pKa = 6.05EE319 pKa = 4.05QLAEE323 pKa = 4.07LEE325 pKa = 4.74IITPLDD331 pKa = 3.47DD332 pKa = 3.61VGASFTNDD340 pKa = 3.6FLPGHH345 pKa = 6.89
MM1 pKa = 7.56LKK3 pKa = 10.22KK4 pKa = 10.37KK5 pKa = 9.51WGTYY9 pKa = 8.36VLAGAFTLMMTACGSSTEE27 pKa = 4.29TEE29 pKa = 4.3STSEE33 pKa = 4.24SVDD36 pKa = 3.49APSDD40 pKa = 3.46DD41 pKa = 4.07PEE43 pKa = 4.54PVEE46 pKa = 4.38EE47 pKa = 4.22TVEE50 pKa = 4.09MTDD53 pKa = 3.47VKK55 pKa = 11.03QVTNWFAQPEE65 pKa = 4.39HH66 pKa = 6.56GGLYY70 pKa = 10.45AALEE74 pKa = 4.02KK75 pKa = 10.96GFYY78 pKa = 9.84EE79 pKa = 4.14EE80 pKa = 5.17NGMNMSIDD88 pKa = 3.22QGGPQVSSIQIVASGQAQFGLAQADD113 pKa = 4.29DD114 pKa = 3.7ILIARR119 pKa = 11.84AEE121 pKa = 4.18GVPIVAIGTIFQTSPQGLLFHH142 pKa = 6.96NSQGITDD149 pKa = 4.34FADD152 pKa = 3.35LQGRR156 pKa = 11.84KK157 pKa = 9.96GIIQPGIGYY166 pKa = 8.88WEE168 pKa = 4.38YY169 pKa = 10.71IKK171 pKa = 10.68EE172 pKa = 4.25FYY174 pKa = 11.01DD175 pKa = 4.03LDD177 pKa = 4.1DD178 pKa = 3.8VQEE181 pKa = 4.13ITFTGQFIDD190 pKa = 4.94FIEE193 pKa = 5.41DD194 pKa = 3.29EE195 pKa = 4.41TSFTQGYY202 pKa = 6.57VTQDD206 pKa = 3.22PFNFEE211 pKa = 4.01QQGLDD216 pKa = 3.11TDD218 pKa = 3.86YY219 pKa = 11.91LLIHH223 pKa = 7.34DD224 pKa = 5.09SGYY227 pKa = 10.28QPYY230 pKa = 10.12ANVMFTTEE238 pKa = 4.38EE239 pKa = 4.34LIEE242 pKa = 4.3NNPEE246 pKa = 3.77LVSAYY251 pKa = 10.54VEE253 pKa = 4.09ASVKK257 pKa = 9.89GWEE260 pKa = 4.55FYY262 pKa = 10.71KK263 pKa = 10.83EE264 pKa = 4.18NYY266 pKa = 10.23DD267 pKa = 3.85EE268 pKa = 4.69INPVIQEE275 pKa = 4.28SNPDD279 pKa = 3.42FPTDD283 pKa = 3.41QLAFTAEE290 pKa = 4.2TLVTYY295 pKa = 10.7AFEE298 pKa = 5.17GDD300 pKa = 3.52AAEE303 pKa = 4.68HH304 pKa = 6.19GFGYY308 pKa = 9.93MSEE311 pKa = 4.3EE312 pKa = 3.89RR313 pKa = 11.84WTTLHH318 pKa = 6.05EE319 pKa = 4.05QLAEE323 pKa = 4.07LEE325 pKa = 4.74IITPLDD331 pKa = 3.47DD332 pKa = 3.61VGASFTNDD340 pKa = 3.6FLPGHH345 pKa = 6.89
Molecular weight: 38.39 kDa
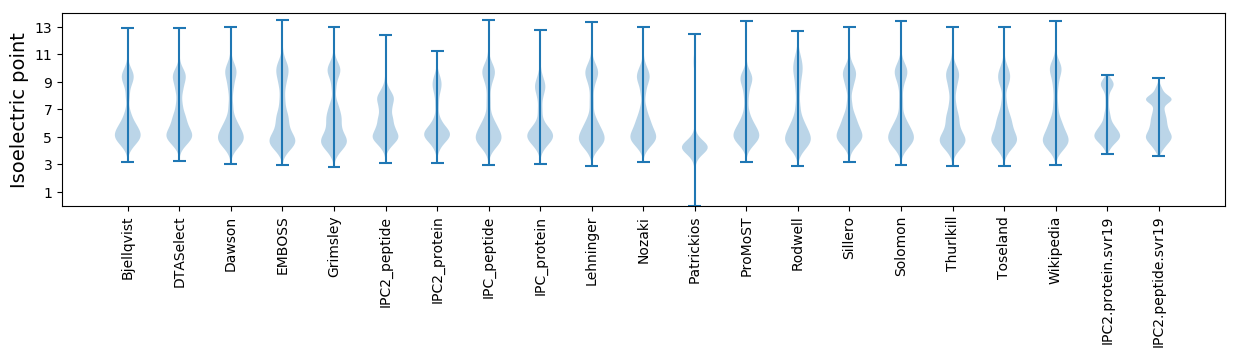
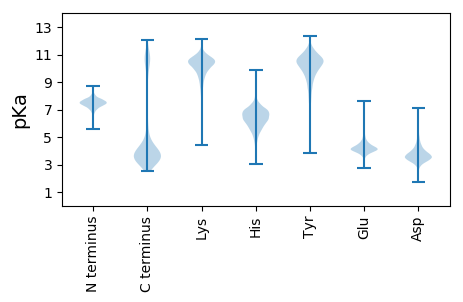
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4Q2N6|W4Q2N6_9BACI Uncharacterized protein OS=Bacillus wakoensis JCM 9140 OX=1236970 GN=JCM9140_2300 PE=4 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1243538 |
37 |
1502 |
271.8 |
30.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.858 ± 0.042 | 0.713 ± 0.012 |
5.075 ± 0.031 | 7.965 ± 0.042 |
4.525 ± 0.03 | 6.704 ± 0.038 |
2.22 ± 0.019 | 7.557 ± 0.03 |
6.446 ± 0.036 | 9.848 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.819 ± 0.017 | 4.204 ± 0.027 |
3.534 ± 0.022 | 4.036 ± 0.029 |
4.201 ± 0.026 | 6.045 ± 0.03 |
5.525 ± 0.024 | 7.385 ± 0.031 |
1.001 ± 0.014 | 3.34 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
