
Rhodotorula graminis (strain WP1)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Pucciniomycotina; Microbotryomycetes; Sporidiobolales; Sporidiobolaceae; Rhodotorula; Rhodotorula graminis
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
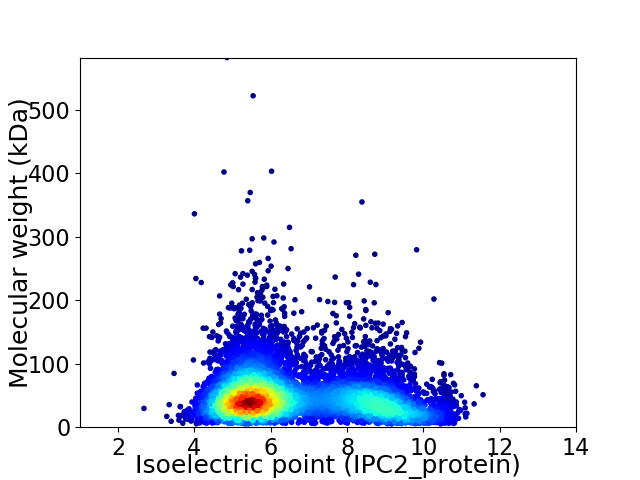
Virtual 2D-PAGE plot for 7228 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A194SAA9|A0A194SAA9_RHOGW Uncharacterized protein (Fragment) OS=Rhodotorula graminis (strain WP1) OX=578459 GN=RHOBADRAFT_65998 PE=4 SV=1
MM1 pKa = 7.72LGRR4 pKa = 11.84ATAAALVVLAGVAQAIVVSSPTATSVWSQSADD36 pKa = 2.73GDD38 pKa = 4.36ARR40 pKa = 11.84IEE42 pKa = 4.08WSLLPATSPPPATQYY57 pKa = 11.15FDD59 pKa = 2.92IYY61 pKa = 10.61LRR63 pKa = 11.84NGVGAMYY70 pKa = 9.65TPSLNLLLATKK81 pKa = 10.12VDD83 pKa = 4.3SLASTYY89 pKa = 11.08LDD91 pKa = 3.22VAATDD96 pKa = 3.78KK97 pKa = 10.69FVPGPGYY104 pKa = 10.13QLFFSDD110 pKa = 4.12PANPATVYY118 pKa = 10.61CDD120 pKa = 3.09SDD122 pKa = 3.72VFAIGSYY129 pKa = 9.7EE130 pKa = 3.95INGPSPTSSSSSDD143 pKa = 3.14VDD145 pKa = 3.63SSTQAEE151 pKa = 4.19ASASSTEE158 pKa = 3.92TRR160 pKa = 11.84AAAAPTSTEE169 pKa = 3.89EE170 pKa = 4.23TTVEE174 pKa = 4.01PSSTEE179 pKa = 3.86TLSYY183 pKa = 10.76EE184 pKa = 4.18PASTDD189 pKa = 3.22STSTSSGMITSVTSSSSPSPAPVSSSSSSAPAGGITAAPAPGGPNEE235 pKa = 4.03QGFNLVPSGASGVRR249 pKa = 11.84AGLSAAAGLSLLAGAGAALLLL270 pKa = 4.39
MM1 pKa = 7.72LGRR4 pKa = 11.84ATAAALVVLAGVAQAIVVSSPTATSVWSQSADD36 pKa = 2.73GDD38 pKa = 4.36ARR40 pKa = 11.84IEE42 pKa = 4.08WSLLPATSPPPATQYY57 pKa = 11.15FDD59 pKa = 2.92IYY61 pKa = 10.61LRR63 pKa = 11.84NGVGAMYY70 pKa = 9.65TPSLNLLLATKK81 pKa = 10.12VDD83 pKa = 4.3SLASTYY89 pKa = 11.08LDD91 pKa = 3.22VAATDD96 pKa = 3.78KK97 pKa = 10.69FVPGPGYY104 pKa = 10.13QLFFSDD110 pKa = 4.12PANPATVYY118 pKa = 10.61CDD120 pKa = 3.09SDD122 pKa = 3.72VFAIGSYY129 pKa = 9.7EE130 pKa = 3.95INGPSPTSSSSSDD143 pKa = 3.14VDD145 pKa = 3.63SSTQAEE151 pKa = 4.19ASASSTEE158 pKa = 3.92TRR160 pKa = 11.84AAAAPTSTEE169 pKa = 3.89EE170 pKa = 4.23TTVEE174 pKa = 4.01PSSTEE179 pKa = 3.86TLSYY183 pKa = 10.76EE184 pKa = 4.18PASTDD189 pKa = 3.22STSTSSGMITSVTSSSSPSPAPVSSSSSSAPAGGITAAPAPGGPNEE235 pKa = 4.03QGFNLVPSGASGVRR249 pKa = 11.84AGLSAAAGLSLLAGAGAALLLL270 pKa = 4.39
Molecular weight: 26.69 kDa
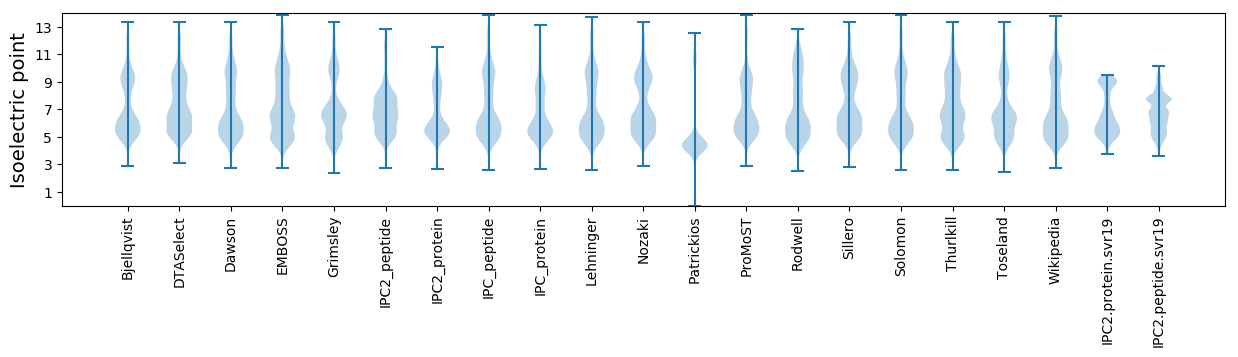
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A194S7E0|A0A194S7E0_RHOGW Uncharacterized protein OS=Rhodotorula graminis (strain WP1) OX=578459 GN=RHOBADRAFT_43836 PE=4 SV=1
RR1 pKa = 7.23RR2 pKa = 11.84TRR4 pKa = 11.84TCTLRR9 pKa = 11.84IAQSTRR15 pKa = 11.84PTSLLPTHH23 pKa = 7.24PSPTSPSSRR32 pKa = 11.84PSRR35 pKa = 11.84TRR37 pKa = 11.84TPSSRR42 pKa = 11.84RR43 pKa = 11.84PTRR46 pKa = 11.84PLPLPLLPRR55 pKa = 11.84TRR57 pKa = 11.84PPGRR61 pKa = 11.84SRR63 pKa = 11.84SATRR67 pKa = 11.84PSRR70 pKa = 11.84RR71 pKa = 11.84TRR73 pKa = 11.84ARR75 pKa = 11.84PRR77 pKa = 11.84SRR79 pKa = 11.84RR80 pKa = 11.84PTRR83 pKa = 11.84PATAAAPSRR92 pKa = 11.84PRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84PARR99 pKa = 11.84PRR101 pKa = 11.84PTSRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84SPSPTRR113 pKa = 11.84LPTATPTASLRR124 pKa = 11.84AASRR128 pKa = 11.84SSSPRR133 pKa = 11.84TAASRR138 pKa = 11.84LLSRR142 pKa = 11.84RR143 pKa = 11.84SATSTAPTSRR153 pKa = 11.84AARR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84PSAASTSRR166 pKa = 11.84AAVRR170 pKa = 11.84GRR172 pKa = 11.84RR173 pKa = 11.84TAPAGPSGRR182 pKa = 11.84RR183 pKa = 11.84GRR185 pKa = 11.84PTASRR190 pKa = 11.84SRR192 pKa = 11.84RR193 pKa = 11.84RR194 pKa = 11.84LSARR198 pKa = 11.84RR199 pKa = 11.84SPRR202 pKa = 11.84TSRR205 pKa = 11.84AASTRR210 pKa = 11.84ATSATSALLARR221 pKa = 11.84APSRR225 pKa = 11.84LTFSRR230 pKa = 11.84TPRR233 pKa = 11.84RR234 pKa = 11.84SRR236 pKa = 11.84SSAAPATEE244 pKa = 4.91GSPSCPTSAVTAAFVATPSRR264 pKa = 11.84PSKK267 pKa = 9.64SRR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84ARR273 pKa = 11.84GRR275 pKa = 11.84LSVRR279 pKa = 11.84HH280 pKa = 5.04RR281 pKa = 11.84AGRR284 pKa = 11.84RR285 pKa = 11.84AAAGLARR292 pKa = 11.84PRR294 pKa = 11.84SCPTRR299 pKa = 11.84VRR301 pKa = 11.84HH302 pKa = 6.55RR303 pKa = 11.84YY304 pKa = 8.82GLSHH308 pKa = 7.64LRR310 pKa = 11.84RR311 pKa = 11.84QGCRR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84GTSPHH322 pKa = 6.08FFFPRR327 pKa = 11.84TLPLVVRR334 pKa = 11.84FF335 pKa = 4.2
RR1 pKa = 7.23RR2 pKa = 11.84TRR4 pKa = 11.84TCTLRR9 pKa = 11.84IAQSTRR15 pKa = 11.84PTSLLPTHH23 pKa = 7.24PSPTSPSSRR32 pKa = 11.84PSRR35 pKa = 11.84TRR37 pKa = 11.84TPSSRR42 pKa = 11.84RR43 pKa = 11.84PTRR46 pKa = 11.84PLPLPLLPRR55 pKa = 11.84TRR57 pKa = 11.84PPGRR61 pKa = 11.84SRR63 pKa = 11.84SATRR67 pKa = 11.84PSRR70 pKa = 11.84RR71 pKa = 11.84TRR73 pKa = 11.84ARR75 pKa = 11.84PRR77 pKa = 11.84SRR79 pKa = 11.84RR80 pKa = 11.84PTRR83 pKa = 11.84PATAAAPSRR92 pKa = 11.84PRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84PARR99 pKa = 11.84PRR101 pKa = 11.84PTSRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84SPSPTRR113 pKa = 11.84LPTATPTASLRR124 pKa = 11.84AASRR128 pKa = 11.84SSSPRR133 pKa = 11.84TAASRR138 pKa = 11.84LLSRR142 pKa = 11.84RR143 pKa = 11.84SATSTAPTSRR153 pKa = 11.84AARR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84PSAASTSRR166 pKa = 11.84AAVRR170 pKa = 11.84GRR172 pKa = 11.84RR173 pKa = 11.84TAPAGPSGRR182 pKa = 11.84RR183 pKa = 11.84GRR185 pKa = 11.84PTASRR190 pKa = 11.84SRR192 pKa = 11.84RR193 pKa = 11.84RR194 pKa = 11.84LSARR198 pKa = 11.84RR199 pKa = 11.84SPRR202 pKa = 11.84TSRR205 pKa = 11.84AASTRR210 pKa = 11.84ATSATSALLARR221 pKa = 11.84APSRR225 pKa = 11.84LTFSRR230 pKa = 11.84TPRR233 pKa = 11.84RR234 pKa = 11.84SRR236 pKa = 11.84SSAAPATEE244 pKa = 4.91GSPSCPTSAVTAAFVATPSRR264 pKa = 11.84PSKK267 pKa = 9.64SRR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84ARR273 pKa = 11.84GRR275 pKa = 11.84LSVRR279 pKa = 11.84HH280 pKa = 5.04RR281 pKa = 11.84AGRR284 pKa = 11.84RR285 pKa = 11.84AAAGLARR292 pKa = 11.84PRR294 pKa = 11.84SCPTRR299 pKa = 11.84VRR301 pKa = 11.84HH302 pKa = 6.55RR303 pKa = 11.84YY304 pKa = 8.82GLSHH308 pKa = 7.64LRR310 pKa = 11.84RR311 pKa = 11.84QGCRR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84GTSPHH322 pKa = 6.08FFFPRR327 pKa = 11.84TLPLVVRR334 pKa = 11.84FF335 pKa = 4.2
Molecular weight: 36.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3636073 |
49 |
5410 |
503.1 |
54.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.082 ± 0.035 | 1.0 ± 0.01 |
5.906 ± 0.024 | 5.689 ± 0.025 |
3.208 ± 0.018 | 7.62 ± 0.03 |
2.423 ± 0.013 | 3.07 ± 0.021 |
3.774 ± 0.029 | 9.423 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.531 ± 0.011 | 2.175 ± 0.016 |
7.219 ± 0.041 | 3.424 ± 0.02 |
7.254 ± 0.038 | 8.736 ± 0.041 |
5.396 ± 0.018 | 6.663 ± 0.025 |
1.262 ± 0.011 | 2.146 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
