
Clostridium puniceum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
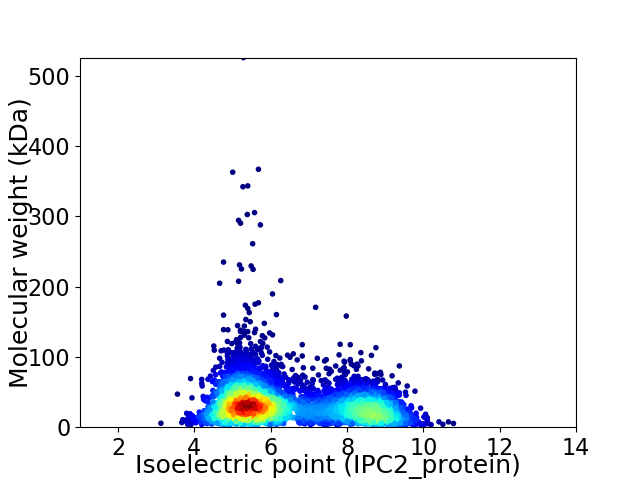
Virtual 2D-PAGE plot for 5300 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S8TWT2|A0A1S8TWT2_9CLOT Bifunctional transcriptional activator/DNA repair enzyme AdaA OS=Clostridium puniceum OX=29367 GN=adaA_1 PE=4 SV=1
MM1 pKa = 7.47NKK3 pKa = 9.88NIKK6 pKa = 10.09RR7 pKa = 11.84IVAMALAISGISAVVPATNFNLFTTKK33 pKa = 10.27AYY35 pKa = 10.48ASSDD39 pKa = 3.62DD40 pKa = 4.73ADD42 pKa = 4.0TLDD45 pKa = 3.96SLEE48 pKa = 5.42LLDD51 pKa = 5.61EE52 pKa = 5.6DD53 pKa = 5.64DD54 pKa = 6.38DD55 pKa = 6.63SIDD58 pKa = 3.82LYY60 pKa = 11.35EE61 pKa = 5.36DD62 pKa = 3.08NDD64 pKa = 3.82YY65 pKa = 11.63EE66 pKa = 4.48EE67 pKa = 5.94EE68 pKa = 4.06IDD70 pKa = 3.63SDD72 pKa = 3.93EE73 pKa = 4.39VEE75 pKa = 4.51EE76 pKa = 4.74GNTYY80 pKa = 10.12YY81 pKa = 11.18AEE83 pKa = 4.31TSSDD87 pKa = 3.2TVHH90 pKa = 7.65IDD92 pKa = 3.75EE93 pKa = 5.75IDD95 pKa = 3.79GADD98 pKa = 3.52QDD100 pKa = 3.66NVRR103 pKa = 11.84IFKK106 pKa = 8.77GTSEE110 pKa = 4.08TAYY113 pKa = 10.2EE114 pKa = 4.12IGDD117 pKa = 4.75DD118 pKa = 3.71ISLSSDD124 pKa = 2.97TTTLKK129 pKa = 10.29VRR131 pKa = 11.84IYY133 pKa = 10.74EE134 pKa = 4.12EE135 pKa = 5.44AYY137 pKa = 10.73DD138 pKa = 5.75DD139 pKa = 4.92YY140 pKa = 11.97DD141 pKa = 3.84DD142 pKa = 5.0SNYY145 pKa = 10.41NEE147 pKa = 4.35YY148 pKa = 9.82EE149 pKa = 4.14VKK151 pKa = 10.55VEE153 pKa = 4.33YY154 pKa = 10.44IGDD157 pKa = 4.26DD158 pKa = 3.77DD159 pKa = 6.45DD160 pKa = 6.68DD161 pKa = 5.56EE162 pKa = 6.24NDD164 pKa = 3.57EE165 pKa = 4.63DD166 pKa = 4.51TLDD169 pKa = 4.05SLKK172 pKa = 10.91LSNADD177 pKa = 2.86GDD179 pKa = 4.46TIKK182 pKa = 10.77LYY184 pKa = 10.93EE185 pKa = 4.79DD186 pKa = 3.91DD187 pKa = 5.43NYY189 pKa = 11.69DD190 pKa = 3.56EE191 pKa = 5.12EE192 pKa = 6.61ADD194 pKa = 3.52GDD196 pKa = 4.3EE197 pKa = 4.36VKK199 pKa = 10.71EE200 pKa = 4.47GYY202 pKa = 8.54TYY204 pKa = 10.78YY205 pKa = 11.21ARR207 pKa = 11.84TSSDD211 pKa = 2.96EE212 pKa = 4.13VEE214 pKa = 4.75IEE216 pKa = 4.22TEE218 pKa = 4.32GPDD221 pKa = 3.2ASYY224 pKa = 11.82VKK226 pKa = 10.37VFKK229 pKa = 10.13STSDD233 pKa = 3.23SAKK236 pKa = 10.76GIDD239 pKa = 3.84PGDD242 pKa = 4.36SISIAGDD249 pKa = 3.01KK250 pKa = 10.72VLTVRR255 pKa = 11.84IYY257 pKa = 10.98SEE259 pKa = 4.84EE260 pKa = 4.19PDD262 pKa = 3.24SDD264 pKa = 3.17ITYY267 pKa = 10.79DD268 pKa = 4.0EE269 pKa = 5.41DD270 pKa = 5.54DD271 pKa = 4.4DD272 pKa = 5.74VIGEE276 pKa = 3.99YY277 pKa = 10.06TIEE280 pKa = 4.57LEE282 pKa = 4.86YY283 pKa = 11.18DD284 pKa = 4.19DD285 pKa = 6.0EE286 pKa = 6.38DD287 pKa = 3.79LTNTSSEE294 pKa = 4.06IVEE297 pKa = 4.46EE298 pKa = 4.21TTEE301 pKa = 4.13TSVTDD306 pKa = 3.27STVITNVVSTIATANQWVQANGKK329 pKa = 6.47WQYY332 pKa = 11.51NDD334 pKa = 5.29AIGNPIKK341 pKa = 10.98NNWFFDD347 pKa = 3.74RR348 pKa = 11.84NLGKK352 pKa = 10.02SYY354 pKa = 10.99YY355 pKa = 10.1LQADD359 pKa = 3.81GNMATGWLSNNSKK372 pKa = 9.67WYY374 pKa = 10.45YY375 pKa = 10.0LGPDD379 pKa = 3.5GAMKK383 pKa = 9.98TGWQLVDD390 pKa = 4.59GIWYY394 pKa = 9.78YY395 pKa = 11.47LDD397 pKa = 3.54SQGVMASNITIDD409 pKa = 4.04GYY411 pKa = 11.84KK412 pKa = 10.4LGANGAWIKK421 pKa = 10.89
MM1 pKa = 7.47NKK3 pKa = 9.88NIKK6 pKa = 10.09RR7 pKa = 11.84IVAMALAISGISAVVPATNFNLFTTKK33 pKa = 10.27AYY35 pKa = 10.48ASSDD39 pKa = 3.62DD40 pKa = 4.73ADD42 pKa = 4.0TLDD45 pKa = 3.96SLEE48 pKa = 5.42LLDD51 pKa = 5.61EE52 pKa = 5.6DD53 pKa = 5.64DD54 pKa = 6.38DD55 pKa = 6.63SIDD58 pKa = 3.82LYY60 pKa = 11.35EE61 pKa = 5.36DD62 pKa = 3.08NDD64 pKa = 3.82YY65 pKa = 11.63EE66 pKa = 4.48EE67 pKa = 5.94EE68 pKa = 4.06IDD70 pKa = 3.63SDD72 pKa = 3.93EE73 pKa = 4.39VEE75 pKa = 4.51EE76 pKa = 4.74GNTYY80 pKa = 10.12YY81 pKa = 11.18AEE83 pKa = 4.31TSSDD87 pKa = 3.2TVHH90 pKa = 7.65IDD92 pKa = 3.75EE93 pKa = 5.75IDD95 pKa = 3.79GADD98 pKa = 3.52QDD100 pKa = 3.66NVRR103 pKa = 11.84IFKK106 pKa = 8.77GTSEE110 pKa = 4.08TAYY113 pKa = 10.2EE114 pKa = 4.12IGDD117 pKa = 4.75DD118 pKa = 3.71ISLSSDD124 pKa = 2.97TTTLKK129 pKa = 10.29VRR131 pKa = 11.84IYY133 pKa = 10.74EE134 pKa = 4.12EE135 pKa = 5.44AYY137 pKa = 10.73DD138 pKa = 5.75DD139 pKa = 4.92YY140 pKa = 11.97DD141 pKa = 3.84DD142 pKa = 5.0SNYY145 pKa = 10.41NEE147 pKa = 4.35YY148 pKa = 9.82EE149 pKa = 4.14VKK151 pKa = 10.55VEE153 pKa = 4.33YY154 pKa = 10.44IGDD157 pKa = 4.26DD158 pKa = 3.77DD159 pKa = 6.45DD160 pKa = 6.68DD161 pKa = 5.56EE162 pKa = 6.24NDD164 pKa = 3.57EE165 pKa = 4.63DD166 pKa = 4.51TLDD169 pKa = 4.05SLKK172 pKa = 10.91LSNADD177 pKa = 2.86GDD179 pKa = 4.46TIKK182 pKa = 10.77LYY184 pKa = 10.93EE185 pKa = 4.79DD186 pKa = 3.91DD187 pKa = 5.43NYY189 pKa = 11.69DD190 pKa = 3.56EE191 pKa = 5.12EE192 pKa = 6.61ADD194 pKa = 3.52GDD196 pKa = 4.3EE197 pKa = 4.36VKK199 pKa = 10.71EE200 pKa = 4.47GYY202 pKa = 8.54TYY204 pKa = 10.78YY205 pKa = 11.21ARR207 pKa = 11.84TSSDD211 pKa = 2.96EE212 pKa = 4.13VEE214 pKa = 4.75IEE216 pKa = 4.22TEE218 pKa = 4.32GPDD221 pKa = 3.2ASYY224 pKa = 11.82VKK226 pKa = 10.37VFKK229 pKa = 10.13STSDD233 pKa = 3.23SAKK236 pKa = 10.76GIDD239 pKa = 3.84PGDD242 pKa = 4.36SISIAGDD249 pKa = 3.01KK250 pKa = 10.72VLTVRR255 pKa = 11.84IYY257 pKa = 10.98SEE259 pKa = 4.84EE260 pKa = 4.19PDD262 pKa = 3.24SDD264 pKa = 3.17ITYY267 pKa = 10.79DD268 pKa = 4.0EE269 pKa = 5.41DD270 pKa = 5.54DD271 pKa = 4.4DD272 pKa = 5.74VIGEE276 pKa = 3.99YY277 pKa = 10.06TIEE280 pKa = 4.57LEE282 pKa = 4.86YY283 pKa = 11.18DD284 pKa = 4.19DD285 pKa = 6.0EE286 pKa = 6.38DD287 pKa = 3.79LTNTSSEE294 pKa = 4.06IVEE297 pKa = 4.46EE298 pKa = 4.21TTEE301 pKa = 4.13TSVTDD306 pKa = 3.27STVITNVVSTIATANQWVQANGKK329 pKa = 6.47WQYY332 pKa = 11.51NDD334 pKa = 5.29AIGNPIKK341 pKa = 10.98NNWFFDD347 pKa = 3.74RR348 pKa = 11.84NLGKK352 pKa = 10.02SYY354 pKa = 10.99YY355 pKa = 10.1LQADD359 pKa = 3.81GNMATGWLSNNSKK372 pKa = 9.67WYY374 pKa = 10.45YY375 pKa = 10.0LGPDD379 pKa = 3.5GAMKK383 pKa = 9.98TGWQLVDD390 pKa = 4.59GIWYY394 pKa = 9.78YY395 pKa = 11.47LDD397 pKa = 3.54SQGVMASNITIDD409 pKa = 4.04GYY411 pKa = 11.84KK412 pKa = 10.4LGANGAWIKK421 pKa = 10.89
Molecular weight: 47.03 kDa
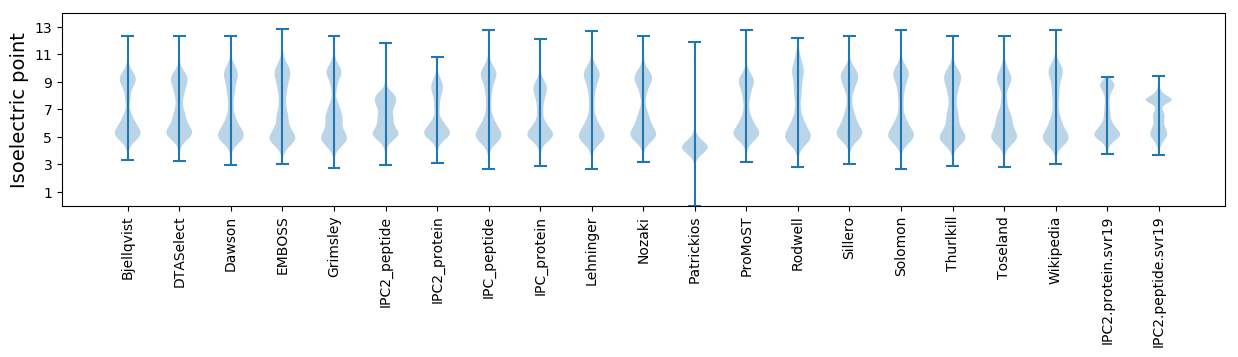
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S8T0W7|A0A1S8T0W7_9CLOT Peptidase_M78 domain-containing protein OS=Clostridium puniceum OX=29367 GN=CLPUN_51070 PE=4 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84EE3 pKa = 3.72EE4 pKa = 4.82GNNSRR9 pKa = 11.84RR10 pKa = 11.84PGGRR14 pKa = 11.84MRR16 pKa = 11.84RR17 pKa = 11.84SRR19 pKa = 11.84KK20 pKa = 9.14KK21 pKa = 9.62VCAFCSDD28 pKa = 2.92KK29 pKa = 11.62SEE31 pKa = 4.79FIDD34 pKa = 3.77YY35 pKa = 11.05KK36 pKa = 11.04DD37 pKa = 3.21INKK40 pKa = 9.06LRR42 pKa = 11.84KK43 pKa = 9.21YY44 pKa = 7.83VTEE47 pKa = 4.22RR48 pKa = 11.84GKK50 pKa = 10.15ILPRR54 pKa = 11.84RR55 pKa = 11.84ISGTCAKK62 pKa = 9.87HH63 pKa = 5.43QRR65 pKa = 11.84EE66 pKa = 4.29LTASIKK72 pKa = 9.88RR73 pKa = 11.84ARR75 pKa = 11.84NIALLPFTTEE85 pKa = 3.59
MM1 pKa = 7.73RR2 pKa = 11.84EE3 pKa = 3.72EE4 pKa = 4.82GNNSRR9 pKa = 11.84RR10 pKa = 11.84PGGRR14 pKa = 11.84MRR16 pKa = 11.84RR17 pKa = 11.84SRR19 pKa = 11.84KK20 pKa = 9.14KK21 pKa = 9.62VCAFCSDD28 pKa = 2.92KK29 pKa = 11.62SEE31 pKa = 4.79FIDD34 pKa = 3.77YY35 pKa = 11.05KK36 pKa = 11.04DD37 pKa = 3.21INKK40 pKa = 9.06LRR42 pKa = 11.84KK43 pKa = 9.21YY44 pKa = 7.83VTEE47 pKa = 4.22RR48 pKa = 11.84GKK50 pKa = 10.15ILPRR54 pKa = 11.84RR55 pKa = 11.84ISGTCAKK62 pKa = 9.87HH63 pKa = 5.43QRR65 pKa = 11.84EE66 pKa = 4.29LTASIKK72 pKa = 9.88RR73 pKa = 11.84ARR75 pKa = 11.84NIALLPFTTEE85 pKa = 3.59
Molecular weight: 9.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1617317 |
29 |
4579 |
305.2 |
34.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.593 ± 0.038 | 1.2 ± 0.014 |
5.5 ± 0.029 | 7.524 ± 0.042 |
4.364 ± 0.025 | 6.284 ± 0.033 |
1.373 ± 0.012 | 9.932 ± 0.042 |
9.027 ± 0.039 | 8.888 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.014 | 6.666 ± 0.029 |
2.701 ± 0.018 | 2.522 ± 0.017 |
3.154 ± 0.019 | 6.46 ± 0.032 |
5.083 ± 0.03 | 6.119 ± 0.03 |
0.818 ± 0.012 | 4.248 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
