
Candidatus Electrothrix marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Candidatus Electrothrix
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
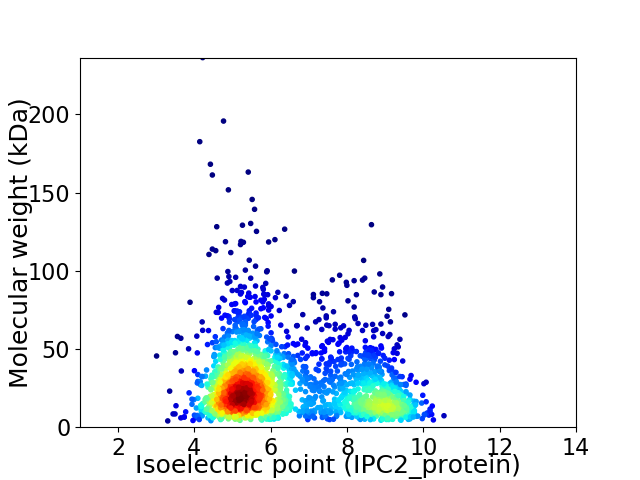
Virtual 2D-PAGE plot for 2161 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
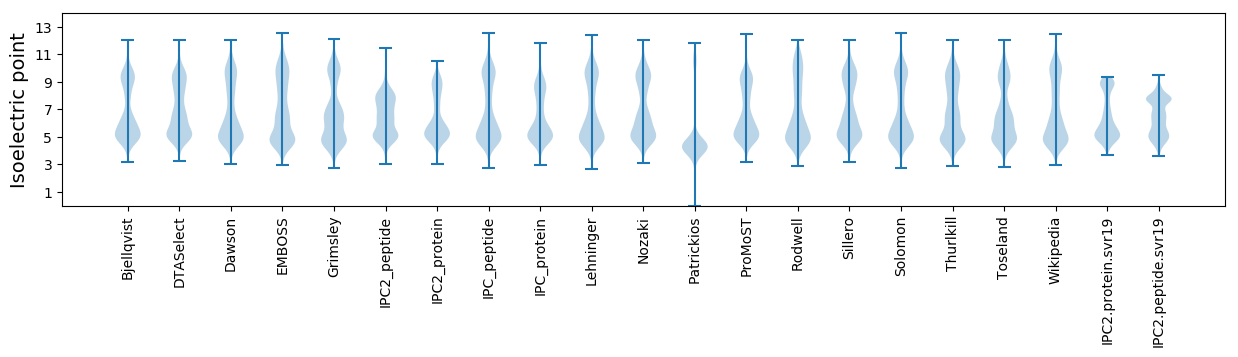
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A444JDS5|A0A444JDS5_9DELT CopG antitoxin of type II toxin-antitoxin system OS=Candidatus Electrothrix marina OX=1859130 GN=VU01_11782 PE=4 SV=1
MM1 pKa = 8.22RR2 pKa = 11.84KK3 pKa = 9.44RR4 pKa = 11.84LWLYY8 pKa = 10.31LVVAEE13 pKa = 4.95ICFCFSGVARR23 pKa = 11.84ADD25 pKa = 3.21IDD27 pKa = 3.66PATVQKK33 pKa = 10.78LFSEE37 pKa = 4.69DD38 pKa = 5.1AGATWLFGYY47 pKa = 9.99SVAVSGNTAVVGSPDD62 pKa = 3.55FSNGGAAYY70 pKa = 10.38VFTRR74 pKa = 11.84SCPEE78 pKa = 3.74STDD81 pKa = 2.74WTVQQKK87 pKa = 10.54LNPPDD92 pKa = 4.23LGFNLNNSGLDD103 pKa = 3.59DD104 pKa = 4.63FGLSVAIDD112 pKa = 3.75KK113 pKa = 8.87DD114 pKa = 3.88TILIGSRR121 pKa = 11.84SWDD124 pKa = 3.2SSLNPGGITVTYY136 pKa = 10.3VFTRR140 pKa = 11.84SEE142 pKa = 4.49GVWGFQQEE150 pKa = 4.05LSIYY154 pKa = 8.03PTVIDD159 pKa = 4.55GKK161 pKa = 8.24WHH163 pKa = 5.35GVKK166 pKa = 10.33NVAVHH171 pKa = 6.34GDD173 pKa = 3.13IAVVVVGGLRR183 pKa = 11.84AVLDD187 pKa = 3.76YY188 pKa = 10.8DD189 pKa = 3.61YY190 pKa = 11.31GIAYY194 pKa = 9.23VFVRR198 pKa = 11.84SAGVWRR204 pKa = 11.84LQQEE208 pKa = 4.02IAVAGSEE215 pKa = 4.23EE216 pKa = 4.53SFGSSVSIDD225 pKa = 3.15GEE227 pKa = 4.63TIVFGAMLDD236 pKa = 3.72DD237 pKa = 5.64DD238 pKa = 4.83NGEE241 pKa = 4.21KK242 pKa = 10.38SGAAYY247 pKa = 10.43VYY249 pKa = 10.19TRR251 pKa = 11.84TGEE254 pKa = 3.67IWSQQQKK261 pKa = 9.78ILCPDD266 pKa = 3.36AGGGNLFGSSVSLSADD282 pKa = 3.26TLLIGASSYY291 pKa = 11.46NEE293 pKa = 3.97NSSGTAYY300 pKa = 10.06IFTRR304 pKa = 11.84FDD306 pKa = 3.88NEE308 pKa = 3.29WSLQQSIAYY317 pKa = 6.85PQLSDD322 pKa = 3.18PTYY325 pKa = 9.66YY326 pKa = 11.33GLFGEE331 pKa = 4.99GEE333 pKa = 4.55VVSLDD338 pKa = 3.36KK339 pKa = 10.71DD340 pKa = 3.7TAVIGGGSSAYY351 pKa = 10.42VFTRR355 pKa = 11.84SGDD358 pKa = 2.9VWTQKK363 pKa = 10.25EE364 pKa = 4.25QLVPDD369 pKa = 4.02YY370 pKa = 11.34GEE372 pKa = 3.72QGNFGFSVSVDD383 pKa = 2.93GGNIVIGSEE392 pKa = 4.06GSVYY396 pKa = 10.36FYY398 pKa = 11.36GQGNQCDD405 pKa = 3.84SDD407 pKa = 4.08GDD409 pKa = 4.9SITDD413 pKa = 3.76DD414 pKa = 3.53QDD416 pKa = 3.53NCPLEE421 pKa = 4.3VNPDD425 pKa = 3.67QIDD428 pKa = 3.55TDD430 pKa = 3.76GDD432 pKa = 4.23NIGDD436 pKa = 3.9ACDD439 pKa = 3.9PRR441 pKa = 11.84PEE443 pKa = 4.29EE444 pKa = 4.12QDD446 pKa = 3.0SDD448 pKa = 3.54TDD450 pKa = 3.58ADD452 pKa = 3.78EE453 pKa = 4.76VYY455 pKa = 10.59DD456 pKa = 4.63YY457 pKa = 11.28IDD459 pKa = 3.75NCSSISNQDD468 pKa = 3.19QKK470 pKa = 11.41DD471 pKa = 3.43TDD473 pKa = 3.79KK474 pKa = 11.74DD475 pKa = 3.87GLGDD479 pKa = 3.71VCDD482 pKa = 4.63SDD484 pKa = 3.92IDD486 pKa = 4.1SDD488 pKa = 4.88GVDD491 pKa = 3.71NEE493 pKa = 4.34QDD495 pKa = 3.58NCPLSANPNQRR506 pKa = 11.84DD507 pKa = 3.55RR508 pKa = 11.84DD509 pKa = 3.74DD510 pKa = 5.49DD511 pKa = 5.05GIGNVCDD518 pKa = 3.9SDD520 pKa = 4.61VVCSFNPTEE529 pKa = 3.88AA530 pKa = 3.86
MM1 pKa = 8.22RR2 pKa = 11.84KK3 pKa = 9.44RR4 pKa = 11.84LWLYY8 pKa = 10.31LVVAEE13 pKa = 4.95ICFCFSGVARR23 pKa = 11.84ADD25 pKa = 3.21IDD27 pKa = 3.66PATVQKK33 pKa = 10.78LFSEE37 pKa = 4.69DD38 pKa = 5.1AGATWLFGYY47 pKa = 9.99SVAVSGNTAVVGSPDD62 pKa = 3.55FSNGGAAYY70 pKa = 10.38VFTRR74 pKa = 11.84SCPEE78 pKa = 3.74STDD81 pKa = 2.74WTVQQKK87 pKa = 10.54LNPPDD92 pKa = 4.23LGFNLNNSGLDD103 pKa = 3.59DD104 pKa = 4.63FGLSVAIDD112 pKa = 3.75KK113 pKa = 8.87DD114 pKa = 3.88TILIGSRR121 pKa = 11.84SWDD124 pKa = 3.2SSLNPGGITVTYY136 pKa = 10.3VFTRR140 pKa = 11.84SEE142 pKa = 4.49GVWGFQQEE150 pKa = 4.05LSIYY154 pKa = 8.03PTVIDD159 pKa = 4.55GKK161 pKa = 8.24WHH163 pKa = 5.35GVKK166 pKa = 10.33NVAVHH171 pKa = 6.34GDD173 pKa = 3.13IAVVVVGGLRR183 pKa = 11.84AVLDD187 pKa = 3.76YY188 pKa = 10.8DD189 pKa = 3.61YY190 pKa = 11.31GIAYY194 pKa = 9.23VFVRR198 pKa = 11.84SAGVWRR204 pKa = 11.84LQQEE208 pKa = 4.02IAVAGSEE215 pKa = 4.23EE216 pKa = 4.53SFGSSVSIDD225 pKa = 3.15GEE227 pKa = 4.63TIVFGAMLDD236 pKa = 3.72DD237 pKa = 5.64DD238 pKa = 4.83NGEE241 pKa = 4.21KK242 pKa = 10.38SGAAYY247 pKa = 10.43VYY249 pKa = 10.19TRR251 pKa = 11.84TGEE254 pKa = 3.67IWSQQQKK261 pKa = 9.78ILCPDD266 pKa = 3.36AGGGNLFGSSVSLSADD282 pKa = 3.26TLLIGASSYY291 pKa = 11.46NEE293 pKa = 3.97NSSGTAYY300 pKa = 10.06IFTRR304 pKa = 11.84FDD306 pKa = 3.88NEE308 pKa = 3.29WSLQQSIAYY317 pKa = 6.85PQLSDD322 pKa = 3.18PTYY325 pKa = 9.66YY326 pKa = 11.33GLFGEE331 pKa = 4.99GEE333 pKa = 4.55VVSLDD338 pKa = 3.36KK339 pKa = 10.71DD340 pKa = 3.7TAVIGGGSSAYY351 pKa = 10.42VFTRR355 pKa = 11.84SGDD358 pKa = 2.9VWTQKK363 pKa = 10.25EE364 pKa = 4.25QLVPDD369 pKa = 4.02YY370 pKa = 11.34GEE372 pKa = 3.72QGNFGFSVSVDD383 pKa = 2.93GGNIVIGSEE392 pKa = 4.06GSVYY396 pKa = 10.36FYY398 pKa = 11.36GQGNQCDD405 pKa = 3.84SDD407 pKa = 4.08GDD409 pKa = 4.9SITDD413 pKa = 3.76DD414 pKa = 3.53QDD416 pKa = 3.53NCPLEE421 pKa = 4.3VNPDD425 pKa = 3.67QIDD428 pKa = 3.55TDD430 pKa = 3.76GDD432 pKa = 4.23NIGDD436 pKa = 3.9ACDD439 pKa = 3.9PRR441 pKa = 11.84PEE443 pKa = 4.29EE444 pKa = 4.12QDD446 pKa = 3.0SDD448 pKa = 3.54TDD450 pKa = 3.58ADD452 pKa = 3.78EE453 pKa = 4.76VYY455 pKa = 10.59DD456 pKa = 4.63YY457 pKa = 11.28IDD459 pKa = 3.75NCSSISNQDD468 pKa = 3.19QKK470 pKa = 11.41DD471 pKa = 3.43TDD473 pKa = 3.79KK474 pKa = 11.74DD475 pKa = 3.87GLGDD479 pKa = 3.71VCDD482 pKa = 4.63SDD484 pKa = 3.92IDD486 pKa = 4.1SDD488 pKa = 4.88GVDD491 pKa = 3.71NEE493 pKa = 4.34QDD495 pKa = 3.58NCPLSANPNQRR506 pKa = 11.84DD507 pKa = 3.55RR508 pKa = 11.84DD509 pKa = 3.74DD510 pKa = 5.49DD511 pKa = 5.05GIGNVCDD518 pKa = 3.9SDD520 pKa = 4.61VVCSFNPTEE529 pKa = 3.88AA530 pKa = 3.86
Molecular weight: 56.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A444JEI0|A0A444JEI0_9DELT ATP-dependent DNA helicase DinG OS=Candidatus Electrothrix marina OX=1859130 GN=VU01_11381 PE=4 SV=1
MM1 pKa = 7.48ARR3 pKa = 11.84LSGVDD8 pKa = 3.52LPRR11 pKa = 11.84NKK13 pKa = 10.46HH14 pKa = 4.79MDD16 pKa = 3.27RR17 pKa = 11.84ALTYY21 pKa = 9.8IYY23 pKa = 10.65GIGLTTARR31 pKa = 11.84AILDD35 pKa = 3.92KK36 pKa = 11.31ADD38 pKa = 5.05LPYY41 pKa = 11.29QMNSDD46 pKa = 4.5DD47 pKa = 5.27LSGDD51 pKa = 3.4DD52 pKa = 3.39VSRR55 pKa = 11.84IRR57 pKa = 11.84KK58 pKa = 9.27IIEE61 pKa = 3.37SDD63 pKa = 3.96YY64 pKa = 11.05VVEE67 pKa = 4.06GDD69 pKa = 3.41RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84EE73 pKa = 3.53VAMDD77 pKa = 3.66IKK79 pKa = 10.84RR80 pKa = 11.84HH81 pKa = 5.85IDD83 pKa = 2.81LGTYY87 pKa = 9.33RR88 pKa = 11.84GRR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 6.51RR93 pKa = 11.84MSLPCRR99 pKa = 11.84GQRR102 pKa = 11.84TKK104 pKa = 10.95TNARR108 pKa = 11.84TRR110 pKa = 11.84KK111 pKa = 8.64GPKK114 pKa = 9.03RR115 pKa = 11.84GAAVRR120 pKa = 11.84KK121 pKa = 9.37KK122 pKa = 10.59
MM1 pKa = 7.48ARR3 pKa = 11.84LSGVDD8 pKa = 3.52LPRR11 pKa = 11.84NKK13 pKa = 10.46HH14 pKa = 4.79MDD16 pKa = 3.27RR17 pKa = 11.84ALTYY21 pKa = 9.8IYY23 pKa = 10.65GIGLTTARR31 pKa = 11.84AILDD35 pKa = 3.92KK36 pKa = 11.31ADD38 pKa = 5.05LPYY41 pKa = 11.29QMNSDD46 pKa = 4.5DD47 pKa = 5.27LSGDD51 pKa = 3.4DD52 pKa = 3.39VSRR55 pKa = 11.84IRR57 pKa = 11.84KK58 pKa = 9.27IIEE61 pKa = 3.37SDD63 pKa = 3.96YY64 pKa = 11.05VVEE67 pKa = 4.06GDD69 pKa = 3.41RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84EE73 pKa = 3.53VAMDD77 pKa = 3.66IKK79 pKa = 10.84RR80 pKa = 11.84HH81 pKa = 5.85IDD83 pKa = 2.81LGTYY87 pKa = 9.33RR88 pKa = 11.84GRR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 6.51RR93 pKa = 11.84MSLPCRR99 pKa = 11.84GQRR102 pKa = 11.84TKK104 pKa = 10.95TNARR108 pKa = 11.84TRR110 pKa = 11.84KK111 pKa = 8.64GPKK114 pKa = 9.03RR115 pKa = 11.84GAAVRR120 pKa = 11.84KK121 pKa = 9.37KK122 pKa = 10.59
Molecular weight: 14.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
594014 |
39 |
2159 |
274.9 |
30.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.297 ± 0.061 | 1.338 ± 0.022 |
5.56 ± 0.04 | 7.014 ± 0.056 |
4.263 ± 0.04 | 7.268 ± 0.052 |
2.089 ± 0.025 | 6.23 ± 0.044 |
5.564 ± 0.053 | 10.455 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.484 ± 0.031 | 3.589 ± 0.039 |
4.29 ± 0.031 | 4.083 ± 0.039 |
5.676 ± 0.052 | 5.92 ± 0.044 |
5.204 ± 0.045 | 6.668 ± 0.043 |
1.067 ± 0.02 | 2.941 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
