
Capybara microvirus Cap3_SP_588
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.43
Get precalculated fractions of proteins
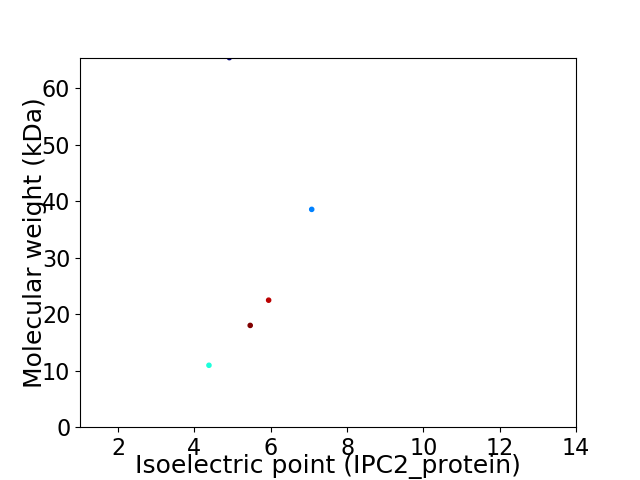
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
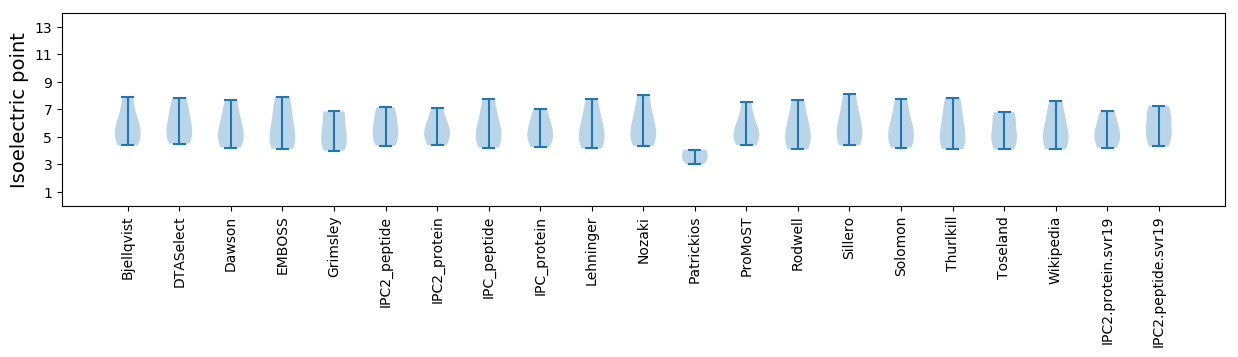
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVV0|A0A4V1FVV0_9VIRU Nonstructural protein OS=Capybara microvirus Cap3_SP_588 OX=2585480 PE=4 SV=1
MM1 pKa = 8.34VEE3 pKa = 4.47MYY5 pKa = 10.57AIYY8 pKa = 10.55DD9 pKa = 3.78PVSEE13 pKa = 4.36SVKK16 pKa = 9.96TPFFIEE22 pKa = 4.55NYY24 pKa = 8.83ASCIRR29 pKa = 11.84AIADD33 pKa = 3.7AYY35 pKa = 10.05NHH37 pKa = 6.33NLFADD42 pKa = 4.47GAKK45 pKa = 10.37DD46 pKa = 4.23VEE48 pKa = 4.35LLSYY52 pKa = 11.08GLIQNDD58 pKa = 3.66GNFYY62 pKa = 10.54PVDD65 pKa = 3.41TDD67 pKa = 3.17IYY69 pKa = 9.27EE70 pKa = 4.54SNGVLVPKK78 pKa = 10.76NFGKK82 pKa = 10.49VFEE85 pKa = 5.09LIKK88 pKa = 11.05DD89 pKa = 3.38EE90 pKa = 5.09SEE92 pKa = 3.98RR93 pKa = 11.84LKK95 pKa = 11.32NGG97 pKa = 3.09
MM1 pKa = 8.34VEE3 pKa = 4.47MYY5 pKa = 10.57AIYY8 pKa = 10.55DD9 pKa = 3.78PVSEE13 pKa = 4.36SVKK16 pKa = 9.96TPFFIEE22 pKa = 4.55NYY24 pKa = 8.83ASCIRR29 pKa = 11.84AIADD33 pKa = 3.7AYY35 pKa = 10.05NHH37 pKa = 6.33NLFADD42 pKa = 4.47GAKK45 pKa = 10.37DD46 pKa = 4.23VEE48 pKa = 4.35LLSYY52 pKa = 11.08GLIQNDD58 pKa = 3.66GNFYY62 pKa = 10.54PVDD65 pKa = 3.41TDD67 pKa = 3.17IYY69 pKa = 9.27EE70 pKa = 4.54SNGVLVPKK78 pKa = 10.76NFGKK82 pKa = 10.49VFEE85 pKa = 5.09LIKK88 pKa = 11.05DD89 pKa = 3.38EE90 pKa = 5.09SEE92 pKa = 3.98RR93 pKa = 11.84LKK95 pKa = 11.32NGG97 pKa = 3.09
Molecular weight: 10.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVV0|A0A4V1FVV0_9VIRU Nonstructural protein OS=Capybara microvirus Cap3_SP_588 OX=2585480 PE=4 SV=1
MM1 pKa = 7.67CLNPKK6 pKa = 9.57KK7 pKa = 10.49AYY9 pKa = 10.23LMNYY13 pKa = 9.57KK14 pKa = 9.94KK15 pKa = 10.7DD16 pKa = 3.53NNKK19 pKa = 9.89NVILFNKK26 pKa = 9.86PIYY29 pKa = 9.79QDD31 pKa = 2.51WHH33 pKa = 5.56KK34 pKa = 10.3IKK36 pKa = 10.73RR37 pKa = 11.84EE38 pKa = 3.6IYY40 pKa = 9.51IPCGICKK47 pKa = 8.91EE48 pKa = 4.47CKK50 pKa = 9.18MSHH53 pKa = 5.88AFDD56 pKa = 3.16WVVRR60 pKa = 11.84CTAEE64 pKa = 4.05SLCYY68 pKa = 9.46EE69 pKa = 3.92NNYY72 pKa = 9.76FVTLTYY78 pKa = 11.19DD79 pKa = 3.23NDD81 pKa = 4.74KK82 pKa = 11.24IPMTPYY88 pKa = 10.57FGDD91 pKa = 4.37DD92 pKa = 4.14DD93 pKa = 4.99SCFMIPTLRR102 pKa = 11.84YY103 pKa = 9.75KK104 pKa = 10.56DD105 pKa = 3.44VQDD108 pKa = 3.47FFKK111 pKa = 10.82RR112 pKa = 11.84LRR114 pKa = 11.84SKK116 pKa = 11.13LDD118 pKa = 3.33YY119 pKa = 10.86NDD121 pKa = 3.76NGNVRR126 pKa = 11.84HH127 pKa = 5.74FTCSEE132 pKa = 3.95YY133 pKa = 11.23GGITQRR139 pKa = 11.84PHH141 pKa = 4.53YY142 pKa = 10.12HH143 pKa = 6.56SILFGINLTDD153 pKa = 3.8LKK155 pKa = 10.91YY156 pKa = 10.85YY157 pKa = 10.8KK158 pKa = 9.38KK159 pKa = 9.24TEE161 pKa = 4.19NGDD164 pKa = 3.19ILYY167 pKa = 9.42TSKK170 pKa = 10.98LLSDD174 pKa = 4.12CWQNGFVIVGEE185 pKa = 4.11VTPEE189 pKa = 3.86SIAYY193 pKa = 8.53VSRR196 pKa = 11.84YY197 pKa = 7.77SLKK200 pKa = 10.11KK201 pKa = 10.34QVDD204 pKa = 3.81EE205 pKa = 4.95TYY207 pKa = 11.12SSYY210 pKa = 11.06HH211 pKa = 6.24LSDD214 pKa = 3.49FRR216 pKa = 11.84EE217 pKa = 4.22SPKK220 pKa = 10.71LYY222 pKa = 8.77MSRR225 pKa = 11.84RR226 pKa = 11.84PGIGAPYY233 pKa = 8.92YY234 pKa = 9.77EE235 pKa = 4.5KK236 pKa = 10.81HH237 pKa = 6.04QIHH240 pKa = 6.42NKK242 pKa = 5.85WQKK245 pKa = 11.03LYY247 pKa = 11.08LNSDD251 pKa = 4.2LKK253 pKa = 11.39DD254 pKa = 3.06IDD256 pKa = 3.72YY257 pKa = 9.45PKK259 pKa = 10.81YY260 pKa = 9.43FLSMLEE266 pKa = 4.07INDD269 pKa = 3.42EE270 pKa = 4.13EE271 pKa = 5.68LYY273 pKa = 10.32IDD275 pKa = 4.38YY276 pKa = 10.04MNNKK280 pKa = 9.99DD281 pKa = 4.45KK282 pKa = 11.21INEE285 pKa = 3.99IKK287 pKa = 10.88ANAYY291 pKa = 10.08YY292 pKa = 9.8PDD294 pKa = 4.48EE295 pKa = 4.26KK296 pKa = 10.45TEE298 pKa = 4.06SYY300 pKa = 10.47RR301 pKa = 11.84LEE303 pKa = 4.1VQARR307 pKa = 11.84KK308 pKa = 10.14YY309 pKa = 10.89KK310 pKa = 10.39FDD312 pKa = 3.47NGYY315 pKa = 10.58LSIRR319 pKa = 11.84NEE321 pKa = 3.97VV322 pKa = 2.98
MM1 pKa = 7.67CLNPKK6 pKa = 9.57KK7 pKa = 10.49AYY9 pKa = 10.23LMNYY13 pKa = 9.57KK14 pKa = 9.94KK15 pKa = 10.7DD16 pKa = 3.53NNKK19 pKa = 9.89NVILFNKK26 pKa = 9.86PIYY29 pKa = 9.79QDD31 pKa = 2.51WHH33 pKa = 5.56KK34 pKa = 10.3IKK36 pKa = 10.73RR37 pKa = 11.84EE38 pKa = 3.6IYY40 pKa = 9.51IPCGICKK47 pKa = 8.91EE48 pKa = 4.47CKK50 pKa = 9.18MSHH53 pKa = 5.88AFDD56 pKa = 3.16WVVRR60 pKa = 11.84CTAEE64 pKa = 4.05SLCYY68 pKa = 9.46EE69 pKa = 3.92NNYY72 pKa = 9.76FVTLTYY78 pKa = 11.19DD79 pKa = 3.23NDD81 pKa = 4.74KK82 pKa = 11.24IPMTPYY88 pKa = 10.57FGDD91 pKa = 4.37DD92 pKa = 4.14DD93 pKa = 4.99SCFMIPTLRR102 pKa = 11.84YY103 pKa = 9.75KK104 pKa = 10.56DD105 pKa = 3.44VQDD108 pKa = 3.47FFKK111 pKa = 10.82RR112 pKa = 11.84LRR114 pKa = 11.84SKK116 pKa = 11.13LDD118 pKa = 3.33YY119 pKa = 10.86NDD121 pKa = 3.76NGNVRR126 pKa = 11.84HH127 pKa = 5.74FTCSEE132 pKa = 3.95YY133 pKa = 11.23GGITQRR139 pKa = 11.84PHH141 pKa = 4.53YY142 pKa = 10.12HH143 pKa = 6.56SILFGINLTDD153 pKa = 3.8LKK155 pKa = 10.91YY156 pKa = 10.85YY157 pKa = 10.8KK158 pKa = 9.38KK159 pKa = 9.24TEE161 pKa = 4.19NGDD164 pKa = 3.19ILYY167 pKa = 9.42TSKK170 pKa = 10.98LLSDD174 pKa = 4.12CWQNGFVIVGEE185 pKa = 4.11VTPEE189 pKa = 3.86SIAYY193 pKa = 8.53VSRR196 pKa = 11.84YY197 pKa = 7.77SLKK200 pKa = 10.11KK201 pKa = 10.34QVDD204 pKa = 3.81EE205 pKa = 4.95TYY207 pKa = 11.12SSYY210 pKa = 11.06HH211 pKa = 6.24LSDD214 pKa = 3.49FRR216 pKa = 11.84EE217 pKa = 4.22SPKK220 pKa = 10.71LYY222 pKa = 8.77MSRR225 pKa = 11.84RR226 pKa = 11.84PGIGAPYY233 pKa = 8.92YY234 pKa = 9.77EE235 pKa = 4.5KK236 pKa = 10.81HH237 pKa = 6.04QIHH240 pKa = 6.42NKK242 pKa = 5.85WQKK245 pKa = 11.03LYY247 pKa = 11.08LNSDD251 pKa = 4.2LKK253 pKa = 11.39DD254 pKa = 3.06IDD256 pKa = 3.72YY257 pKa = 9.45PKK259 pKa = 10.81YY260 pKa = 9.43FLSMLEE266 pKa = 4.07INDD269 pKa = 3.42EE270 pKa = 4.13EE271 pKa = 5.68LYY273 pKa = 10.32IDD275 pKa = 4.38YY276 pKa = 10.04MNNKK280 pKa = 9.99DD281 pKa = 4.45KK282 pKa = 11.21INEE285 pKa = 3.99IKK287 pKa = 10.88ANAYY291 pKa = 10.08YY292 pKa = 9.8PDD294 pKa = 4.48EE295 pKa = 4.26KK296 pKa = 10.45TEE298 pKa = 4.06SYY300 pKa = 10.47RR301 pKa = 11.84LEE303 pKa = 4.1VQARR307 pKa = 11.84KK308 pKa = 10.14YY309 pKa = 10.89KK310 pKa = 10.39FDD312 pKa = 3.47NGYY315 pKa = 10.58LSIRR319 pKa = 11.84NEE321 pKa = 3.97VV322 pKa = 2.98
Molecular weight: 38.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369 |
97 |
579 |
273.8 |
31.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.501 ± 1.557 | 0.95 ± 0.538 |
6.793 ± 0.465 | 5.186 ± 0.638 |
5.186 ± 0.522 | 6.355 ± 0.773 |
1.899 ± 0.288 | 6.209 ± 0.325 |
5.698 ± 1.691 | 6.501 ± 0.43 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.899 ± 0.235 | 7.889 ± 0.724 |
4.602 ± 0.825 | 3.725 ± 0.808 |
4.018 ± 0.593 | 8.4 ± 0.778 |
4.894 ± 0.572 | 5.478 ± 0.801 |
1.242 ± 0.328 | 6.574 ± 0.975 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
