
Salinivirga cyanobacteriivorans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Salinivirgaceae; Salinivirga
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
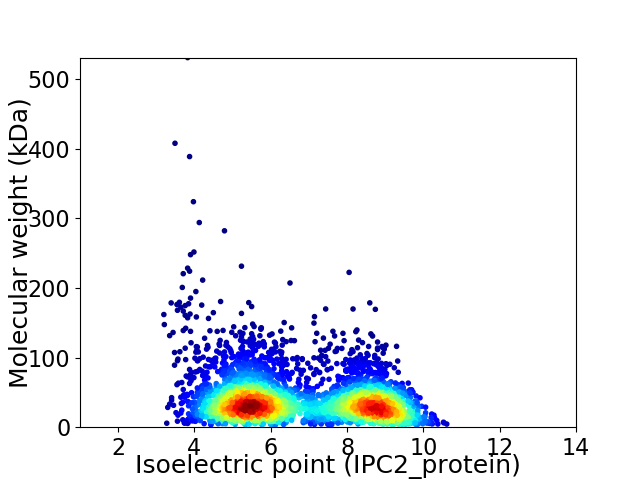
Virtual 2D-PAGE plot for 3717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2HXA5|A0A0S2HXA5_9BACT Dihydroorotate dehydrogenase A (Fumarate) OS=Salinivirga cyanobacteriivorans OX=1307839 GN=pyrDA_1 PE=4 SV=1
MM1 pKa = 7.52LGKK4 pKa = 10.18KK5 pKa = 9.25IKK7 pKa = 10.66YY8 pKa = 9.56LIQILAATFLMLWVTSSCVDD28 pKa = 4.4DD29 pKa = 5.84NFDD32 pKa = 4.07LDD34 pKa = 3.71KK35 pKa = 11.46LSTHH39 pKa = 7.19LEE41 pKa = 4.24NEE43 pKa = 4.58SQWNFPVVSTTFAMEE58 pKa = 5.46DD59 pKa = 3.06ILNLVDD65 pKa = 3.94STGLIYY71 pKa = 10.56TDD73 pKa = 5.3DD74 pKa = 3.85EE75 pKa = 4.76GLVYY79 pKa = 10.61LVYY82 pKa = 10.89SDD84 pKa = 3.7TAYY87 pKa = 9.67TAIAEE92 pKa = 4.24EE93 pKa = 4.83EE94 pKa = 4.38IVFPDD99 pKa = 3.39QNYY102 pKa = 8.24DD103 pKa = 3.77TIFNEE108 pKa = 4.75QDD110 pKa = 3.39YY111 pKa = 11.4QDD113 pKa = 4.34AGGFNNDD120 pKa = 3.04TVKK123 pKa = 10.22MEE125 pKa = 4.29KK126 pKa = 10.55ADD128 pKa = 3.63VNYY131 pKa = 10.62LFVASAEE138 pKa = 4.11NQLLDD143 pKa = 3.74SVKK146 pKa = 10.57VKK148 pKa = 10.92SSLLDD153 pKa = 3.17IDD155 pKa = 4.44VNSSFEE161 pKa = 4.2FEE163 pKa = 4.58GEE165 pKa = 4.02LTLTFPTLRR174 pKa = 11.84KK175 pKa = 9.97NGQLYY180 pKa = 10.29QKK182 pKa = 10.87VIDD185 pKa = 4.17INNASGNFSYY195 pKa = 11.08SEE197 pKa = 4.2TFDD200 pKa = 5.42DD201 pKa = 5.01LDD203 pKa = 5.74GYY205 pKa = 10.61MLDD208 pKa = 3.67FTEE211 pKa = 5.47FAIPDD216 pKa = 3.81SNVFFITYY224 pKa = 8.66EE225 pKa = 3.84LEE227 pKa = 4.33YY228 pKa = 11.16YY229 pKa = 10.54EE230 pKa = 4.47STGSGNVNADD240 pKa = 4.61DD241 pKa = 4.0ICDD244 pKa = 3.41IDD246 pKa = 3.87VTFRR250 pKa = 11.84DD251 pKa = 3.15IEE253 pKa = 4.0YY254 pKa = 9.8DD255 pKa = 4.06YY256 pKa = 11.53VWGYY260 pKa = 10.55IGNQLINIPEE270 pKa = 3.91NNIYY274 pKa = 10.11IDD276 pKa = 3.81LFNEE280 pKa = 4.33FEE282 pKa = 4.13YY283 pKa = 11.61GEE285 pKa = 4.17FQLYY289 pKa = 10.35DD290 pKa = 3.55PRR292 pKa = 11.84LKK294 pKa = 10.7INIFNSFGIPVGVFMNTLQLKK315 pKa = 10.32VDD317 pKa = 4.08GTWEE321 pKa = 4.24DD322 pKa = 3.6VTGSDD327 pKa = 4.87IPTEE331 pKa = 4.1ANPWHH336 pKa = 6.5IPIPEE341 pKa = 4.17SNYY344 pKa = 10.2PEE346 pKa = 4.6IPVADD351 pKa = 4.08TTVMITGQGSNIDD364 pKa = 3.88SLVSRR369 pKa = 11.84LPNNLRR375 pKa = 11.84FAEE378 pKa = 4.27TVEE381 pKa = 4.1INPDD385 pKa = 3.17EE386 pKa = 4.29TGMRR390 pKa = 11.84NNFMAAEE397 pKa = 4.17SRR399 pKa = 11.84VEE401 pKa = 4.12AIVDD405 pKa = 3.76FEE407 pKa = 4.13IPIWGRR413 pKa = 11.84TSGITYY419 pKa = 10.32SDD421 pKa = 4.38TIDD424 pKa = 3.81MDD426 pKa = 4.12LGDD429 pKa = 4.81IGDD432 pKa = 3.9EE433 pKa = 4.05NDD435 pKa = 3.33YY436 pKa = 11.54LDD438 pKa = 4.11YY439 pKa = 11.6LNLKK443 pKa = 8.24LTIGNGFPHH452 pKa = 7.0NIGVKK457 pKa = 10.0GYY459 pKa = 11.14LMDD462 pKa = 3.45STYY465 pKa = 11.11NIVDD469 pKa = 4.82SIPNSNDD476 pKa = 3.08NPNDD480 pKa = 3.31TLIIVEE486 pKa = 4.37SGIIGADD493 pKa = 3.21SLINQEE499 pKa = 3.88TGKK502 pKa = 8.28TVVSSTITYY511 pKa = 9.23EE512 pKa = 4.0NEE514 pKa = 4.05SVDD517 pKa = 3.98VIDD520 pKa = 3.76NVKK523 pKa = 10.86YY524 pKa = 10.11MLLKK528 pKa = 10.51VKK530 pKa = 10.41FMTTDD535 pKa = 3.0GNAPDD540 pKa = 3.51APYY543 pKa = 11.19VKK545 pKa = 10.42FYY547 pKa = 11.25DD548 pKa = 4.3FYY550 pKa = 11.9EE551 pKa = 3.89MDD553 pKa = 3.68FDD555 pKa = 4.94VNVDD559 pKa = 4.23FKK561 pKa = 11.79LDD563 pKa = 3.59VNEE566 pKa = 4.12NLL568 pKa = 4.22
MM1 pKa = 7.52LGKK4 pKa = 10.18KK5 pKa = 9.25IKK7 pKa = 10.66YY8 pKa = 9.56LIQILAATFLMLWVTSSCVDD28 pKa = 4.4DD29 pKa = 5.84NFDD32 pKa = 4.07LDD34 pKa = 3.71KK35 pKa = 11.46LSTHH39 pKa = 7.19LEE41 pKa = 4.24NEE43 pKa = 4.58SQWNFPVVSTTFAMEE58 pKa = 5.46DD59 pKa = 3.06ILNLVDD65 pKa = 3.94STGLIYY71 pKa = 10.56TDD73 pKa = 5.3DD74 pKa = 3.85EE75 pKa = 4.76GLVYY79 pKa = 10.61LVYY82 pKa = 10.89SDD84 pKa = 3.7TAYY87 pKa = 9.67TAIAEE92 pKa = 4.24EE93 pKa = 4.83EE94 pKa = 4.38IVFPDD99 pKa = 3.39QNYY102 pKa = 8.24DD103 pKa = 3.77TIFNEE108 pKa = 4.75QDD110 pKa = 3.39YY111 pKa = 11.4QDD113 pKa = 4.34AGGFNNDD120 pKa = 3.04TVKK123 pKa = 10.22MEE125 pKa = 4.29KK126 pKa = 10.55ADD128 pKa = 3.63VNYY131 pKa = 10.62LFVASAEE138 pKa = 4.11NQLLDD143 pKa = 3.74SVKK146 pKa = 10.57VKK148 pKa = 10.92SSLLDD153 pKa = 3.17IDD155 pKa = 4.44VNSSFEE161 pKa = 4.2FEE163 pKa = 4.58GEE165 pKa = 4.02LTLTFPTLRR174 pKa = 11.84KK175 pKa = 9.97NGQLYY180 pKa = 10.29QKK182 pKa = 10.87VIDD185 pKa = 4.17INNASGNFSYY195 pKa = 11.08SEE197 pKa = 4.2TFDD200 pKa = 5.42DD201 pKa = 5.01LDD203 pKa = 5.74GYY205 pKa = 10.61MLDD208 pKa = 3.67FTEE211 pKa = 5.47FAIPDD216 pKa = 3.81SNVFFITYY224 pKa = 8.66EE225 pKa = 3.84LEE227 pKa = 4.33YY228 pKa = 11.16YY229 pKa = 10.54EE230 pKa = 4.47STGSGNVNADD240 pKa = 4.61DD241 pKa = 4.0ICDD244 pKa = 3.41IDD246 pKa = 3.87VTFRR250 pKa = 11.84DD251 pKa = 3.15IEE253 pKa = 4.0YY254 pKa = 9.8DD255 pKa = 4.06YY256 pKa = 11.53VWGYY260 pKa = 10.55IGNQLINIPEE270 pKa = 3.91NNIYY274 pKa = 10.11IDD276 pKa = 3.81LFNEE280 pKa = 4.33FEE282 pKa = 4.13YY283 pKa = 11.61GEE285 pKa = 4.17FQLYY289 pKa = 10.35DD290 pKa = 3.55PRR292 pKa = 11.84LKK294 pKa = 10.7INIFNSFGIPVGVFMNTLQLKK315 pKa = 10.32VDD317 pKa = 4.08GTWEE321 pKa = 4.24DD322 pKa = 3.6VTGSDD327 pKa = 4.87IPTEE331 pKa = 4.1ANPWHH336 pKa = 6.5IPIPEE341 pKa = 4.17SNYY344 pKa = 10.2PEE346 pKa = 4.6IPVADD351 pKa = 4.08TTVMITGQGSNIDD364 pKa = 3.88SLVSRR369 pKa = 11.84LPNNLRR375 pKa = 11.84FAEE378 pKa = 4.27TVEE381 pKa = 4.1INPDD385 pKa = 3.17EE386 pKa = 4.29TGMRR390 pKa = 11.84NNFMAAEE397 pKa = 4.17SRR399 pKa = 11.84VEE401 pKa = 4.12AIVDD405 pKa = 3.76FEE407 pKa = 4.13IPIWGRR413 pKa = 11.84TSGITYY419 pKa = 10.32SDD421 pKa = 4.38TIDD424 pKa = 3.81MDD426 pKa = 4.12LGDD429 pKa = 4.81IGDD432 pKa = 3.9EE433 pKa = 4.05NDD435 pKa = 3.33YY436 pKa = 11.54LDD438 pKa = 4.11YY439 pKa = 11.6LNLKK443 pKa = 8.24LTIGNGFPHH452 pKa = 7.0NIGVKK457 pKa = 10.0GYY459 pKa = 11.14LMDD462 pKa = 3.45STYY465 pKa = 11.11NIVDD469 pKa = 4.82SIPNSNDD476 pKa = 3.08NPNDD480 pKa = 3.31TLIIVEE486 pKa = 4.37SGIIGADD493 pKa = 3.21SLINQEE499 pKa = 3.88TGKK502 pKa = 8.28TVVSSTITYY511 pKa = 9.23EE512 pKa = 4.0NEE514 pKa = 4.05SVDD517 pKa = 3.98VIDD520 pKa = 3.76NVKK523 pKa = 10.86YY524 pKa = 10.11MLLKK528 pKa = 10.51VKK530 pKa = 10.41FMTTDD535 pKa = 3.0GNAPDD540 pKa = 3.51APYY543 pKa = 11.19VKK545 pKa = 10.42FYY547 pKa = 11.25DD548 pKa = 4.3FYY550 pKa = 11.9EE551 pKa = 3.89MDD553 pKa = 3.68FDD555 pKa = 4.94VNVDD559 pKa = 4.23FKK561 pKa = 11.79LDD563 pKa = 3.59VNEE566 pKa = 4.12NLL568 pKa = 4.22
Molecular weight: 64.22 kDa
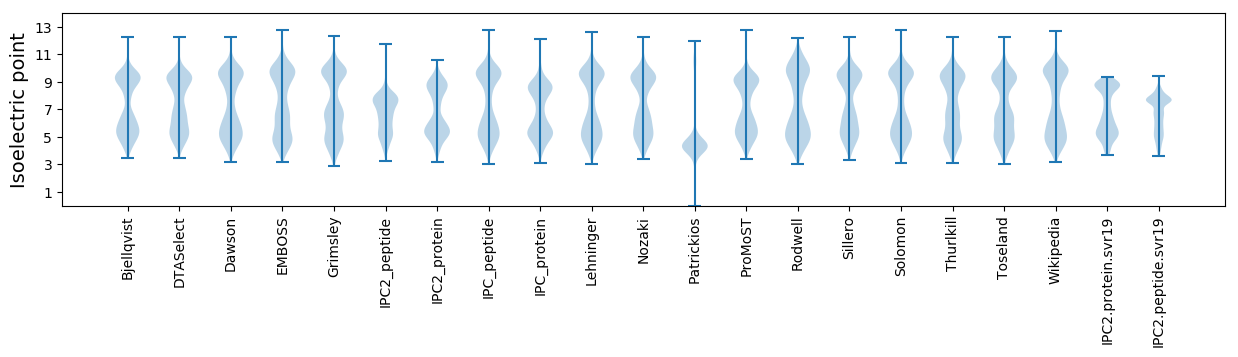
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2I2H8|A0A0S2I2H8_9BACT TPR repeat-containing protein YrrB OS=Salinivirga cyanobacteriivorans OX=1307839 GN=yrrB_6 PE=3 SV=1
MM1 pKa = 7.43LNDD4 pKa = 4.15AKK6 pKa = 10.68QPHH9 pKa = 5.9WGKK12 pKa = 10.17RR13 pKa = 11.84RR14 pKa = 11.84CCFANYY20 pKa = 7.66MLPLIEE26 pKa = 4.07MFSRR30 pKa = 11.84HH31 pKa = 5.55EE32 pKa = 3.83RR33 pKa = 11.84HH34 pKa = 5.84NDD36 pKa = 2.61NRR38 pKa = 11.84QRR40 pKa = 11.84TPRR43 pKa = 11.84VPTLRR48 pKa = 11.84TLRR51 pKa = 11.84NLSGRR56 pKa = 11.84CVKK59 pKa = 10.38IIFEE63 pKa = 4.3GVIGRR68 pKa = 11.84RR69 pKa = 11.84YY70 pKa = 10.54NEE72 pKa = 3.6ANYY75 pKa = 10.89
MM1 pKa = 7.43LNDD4 pKa = 4.15AKK6 pKa = 10.68QPHH9 pKa = 5.9WGKK12 pKa = 10.17RR13 pKa = 11.84RR14 pKa = 11.84CCFANYY20 pKa = 7.66MLPLIEE26 pKa = 4.07MFSRR30 pKa = 11.84HH31 pKa = 5.55EE32 pKa = 3.83RR33 pKa = 11.84HH34 pKa = 5.84NDD36 pKa = 2.61NRR38 pKa = 11.84QRR40 pKa = 11.84TPRR43 pKa = 11.84VPTLRR48 pKa = 11.84TLRR51 pKa = 11.84NLSGRR56 pKa = 11.84CVKK59 pKa = 10.38IIFEE63 pKa = 4.3GVIGRR68 pKa = 11.84RR69 pKa = 11.84YY70 pKa = 10.54NEE72 pKa = 3.6ANYY75 pKa = 10.89
Molecular weight: 9.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369904 |
29 |
4899 |
368.6 |
41.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.582 ± 0.04 | 0.681 ± 0.013 |
5.759 ± 0.039 | 6.721 ± 0.041 |
4.971 ± 0.031 | 6.427 ± 0.05 |
2.046 ± 0.017 | 7.754 ± 0.035 |
7.339 ± 0.072 | 8.97 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.02 | 5.873 ± 0.039 |
3.53 ± 0.024 | 3.817 ± 0.027 |
3.975 ± 0.032 | 6.069 ± 0.035 |
5.586 ± 0.051 | 6.043 ± 0.033 |
1.095 ± 0.015 | 4.358 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
