
Camel associated porprismacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
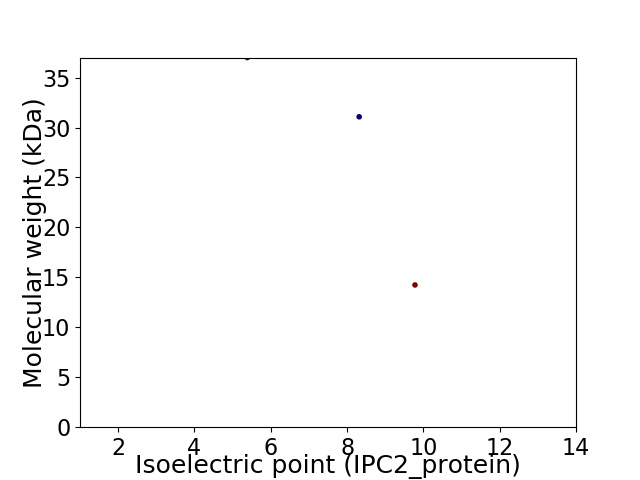
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
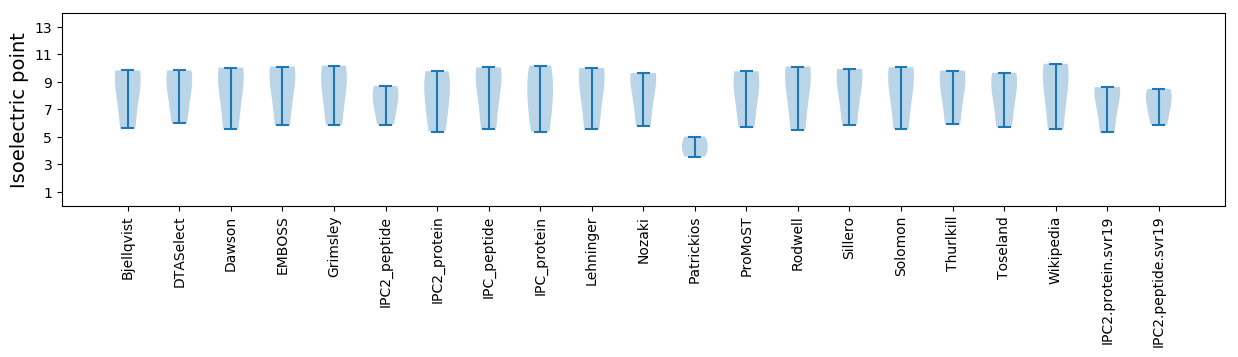

Protein with the lowest isoelectric point:
>tr|A0A0A1ENV3|A0A0A1ENV3_9VIRU Uncharacterized protein OS=Camel associated porprismacovirus 1 OX=2170105 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.01SQSITVSIVEE11 pKa = 4.58TYY13 pKa = 10.93DD14 pKa = 3.91LSTTEE19 pKa = 4.0NRR21 pKa = 11.84MGLIAIKK28 pKa = 8.7TPSMTAVNKK37 pKa = 10.05RR38 pKa = 11.84YY39 pKa = 9.51PGFIRR44 pKa = 11.84NFKK47 pKa = 9.35FLHH50 pKa = 5.86VNSCDD55 pKa = 3.62VVLSCASTLPTDD67 pKa = 4.08PLQIGTTAGKK77 pKa = 9.78VAPQDD82 pKa = 3.32MFNPLLYY89 pKa = 10.4KK90 pKa = 10.52AVSNDD95 pKa = 2.65SWNGLLSRR103 pKa = 11.84IYY105 pKa = 10.41SPTGASVSLNDD116 pKa = 4.69DD117 pKa = 3.18VRR119 pKa = 11.84HH120 pKa = 5.79FADD123 pKa = 4.55AFPDD127 pKa = 3.57ATSTEE132 pKa = 4.45SVRR135 pKa = 11.84AYY137 pKa = 10.75YY138 pKa = 11.29AMLADD143 pKa = 4.05PSFKK147 pKa = 10.1KK148 pKa = 10.69AHH150 pKa = 5.72IQAGLSMRR158 pKa = 11.84GLRR161 pKa = 11.84PLVYY165 pKa = 10.02HH166 pKa = 6.84LLSGSGTNTEE176 pKa = 4.18LTGAGQPTAVGQAYY190 pKa = 9.69KK191 pKa = 10.88NYY193 pKa = 10.08VDD195 pKa = 5.29DD196 pKa = 6.0SGDD199 pKa = 3.31AYY201 pKa = 10.95QRR203 pKa = 11.84ASNNLYY209 pKa = 10.78SGTTLFKK216 pKa = 10.71GRR218 pKa = 11.84PVPMPSVQTTPAVVHH233 pKa = 6.49PSDD236 pKa = 3.71GTVTQIDD243 pKa = 4.41YY244 pKa = 11.15NVVLGGIPSTYY255 pKa = 9.05VCAIVVPPHH264 pKa = 6.31SLNTTYY270 pKa = 10.78FRR272 pKa = 11.84MTVKK276 pKa = 10.31WSVTFHH282 pKa = 5.79TVASEE287 pKa = 3.84MSKK290 pKa = 11.28ALMPGMSDD298 pKa = 3.5LGRR301 pKa = 11.84YY302 pKa = 8.77AYY304 pKa = 7.75YY305 pKa = 10.33TSEE308 pKa = 4.13TSKK311 pKa = 11.1LEE313 pKa = 3.85PVPIEE318 pKa = 4.5DD319 pKa = 3.99VSDD322 pKa = 3.56TAVTGDD328 pKa = 3.48TVDD331 pKa = 3.54ADD333 pKa = 4.26GVSLDD338 pKa = 4.39LVMEE342 pKa = 4.57KK343 pKa = 10.8
MM1 pKa = 7.01SQSITVSIVEE11 pKa = 4.58TYY13 pKa = 10.93DD14 pKa = 3.91LSTTEE19 pKa = 4.0NRR21 pKa = 11.84MGLIAIKK28 pKa = 8.7TPSMTAVNKK37 pKa = 10.05RR38 pKa = 11.84YY39 pKa = 9.51PGFIRR44 pKa = 11.84NFKK47 pKa = 9.35FLHH50 pKa = 5.86VNSCDD55 pKa = 3.62VVLSCASTLPTDD67 pKa = 4.08PLQIGTTAGKK77 pKa = 9.78VAPQDD82 pKa = 3.32MFNPLLYY89 pKa = 10.4KK90 pKa = 10.52AVSNDD95 pKa = 2.65SWNGLLSRR103 pKa = 11.84IYY105 pKa = 10.41SPTGASVSLNDD116 pKa = 4.69DD117 pKa = 3.18VRR119 pKa = 11.84HH120 pKa = 5.79FADD123 pKa = 4.55AFPDD127 pKa = 3.57ATSTEE132 pKa = 4.45SVRR135 pKa = 11.84AYY137 pKa = 10.75YY138 pKa = 11.29AMLADD143 pKa = 4.05PSFKK147 pKa = 10.1KK148 pKa = 10.69AHH150 pKa = 5.72IQAGLSMRR158 pKa = 11.84GLRR161 pKa = 11.84PLVYY165 pKa = 10.02HH166 pKa = 6.84LLSGSGTNTEE176 pKa = 4.18LTGAGQPTAVGQAYY190 pKa = 9.69KK191 pKa = 10.88NYY193 pKa = 10.08VDD195 pKa = 5.29DD196 pKa = 6.0SGDD199 pKa = 3.31AYY201 pKa = 10.95QRR203 pKa = 11.84ASNNLYY209 pKa = 10.78SGTTLFKK216 pKa = 10.71GRR218 pKa = 11.84PVPMPSVQTTPAVVHH233 pKa = 6.49PSDD236 pKa = 3.71GTVTQIDD243 pKa = 4.41YY244 pKa = 11.15NVVLGGIPSTYY255 pKa = 9.05VCAIVVPPHH264 pKa = 6.31SLNTTYY270 pKa = 10.78FRR272 pKa = 11.84MTVKK276 pKa = 10.31WSVTFHH282 pKa = 5.79TVASEE287 pKa = 3.84MSKK290 pKa = 11.28ALMPGMSDD298 pKa = 3.5LGRR301 pKa = 11.84YY302 pKa = 8.77AYY304 pKa = 7.75YY305 pKa = 10.33TSEE308 pKa = 4.13TSKK311 pKa = 11.1LEE313 pKa = 3.85PVPIEE318 pKa = 4.5DD319 pKa = 3.99VSDD322 pKa = 3.56TAVTGDD328 pKa = 3.48TVDD331 pKa = 3.54ADD333 pKa = 4.26GVSLDD338 pKa = 4.39LVMEE342 pKa = 4.57KK343 pKa = 10.8
Molecular weight: 37.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1EJ70|A0A0A1EJ70_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 1 OX=2170105 GN=rep PE=4 SV=1
MM1 pKa = 7.27YY2 pKa = 9.05PILPLLVILSMPMVFLSILSWRR24 pKa = 11.84NDD26 pKa = 3.08MASYY30 pKa = 9.97RR31 pKa = 11.84QYY33 pKa = 11.09RR34 pKa = 11.84HH35 pKa = 5.42QSSSNFGRR43 pKa = 11.84PSSMRR48 pKa = 11.84FDD50 pKa = 4.48LDD52 pKa = 2.9PRR54 pKa = 11.84TRR56 pKa = 11.84YY57 pKa = 9.38EE58 pKa = 3.79LSKK61 pKa = 11.27VPFIGDD67 pKa = 3.63AFRR70 pKa = 11.84YY71 pKa = 8.48MDD73 pKa = 3.13QTKK76 pKa = 10.65YY77 pKa = 10.85YY78 pKa = 10.29NDD80 pKa = 3.51YY81 pKa = 10.21LASRR85 pKa = 11.84GMTWNDD91 pKa = 2.63VKK93 pKa = 11.36YY94 pKa = 9.47PALLSGSGAYY104 pKa = 9.31GVAASGYY111 pKa = 8.17RR112 pKa = 11.84VSKK115 pKa = 11.04NFLKK119 pKa = 10.81LYY121 pKa = 10.13RR122 pKa = 4.74
MM1 pKa = 7.27YY2 pKa = 9.05PILPLLVILSMPMVFLSILSWRR24 pKa = 11.84NDD26 pKa = 3.08MASYY30 pKa = 9.97RR31 pKa = 11.84QYY33 pKa = 11.09RR34 pKa = 11.84HH35 pKa = 5.42QSSSNFGRR43 pKa = 11.84PSSMRR48 pKa = 11.84FDD50 pKa = 4.48LDD52 pKa = 2.9PRR54 pKa = 11.84TRR56 pKa = 11.84YY57 pKa = 9.38EE58 pKa = 3.79LSKK61 pKa = 11.27VPFIGDD67 pKa = 3.63AFRR70 pKa = 11.84YY71 pKa = 8.48MDD73 pKa = 3.13QTKK76 pKa = 10.65YY77 pKa = 10.85YY78 pKa = 10.29NDD80 pKa = 3.51YY81 pKa = 10.21LASRR85 pKa = 11.84GMTWNDD91 pKa = 2.63VKK93 pKa = 11.36YY94 pKa = 9.47PALLSGSGAYY104 pKa = 9.31GVAASGYY111 pKa = 8.17RR112 pKa = 11.84VSKK115 pKa = 11.04NFLKK119 pKa = 10.81LYY121 pKa = 10.13RR122 pKa = 4.74
Molecular weight: 14.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
732 |
122 |
343 |
244.0 |
27.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.557 ± 0.808 | 1.23 ± 0.47 |
6.148 ± 0.163 | 4.372 ± 1.766 |
3.552 ± 0.417 | 6.694 ± 0.242 |
2.049 ± 0.318 | 4.781 ± 1.082 |
5.601 ± 1.42 | 7.923 ± 0.687 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.279 ± 0.708 | 3.689 ± 0.323 |
4.918 ± 1.107 | 2.732 ± 0.121 |
6.284 ± 1.678 | 8.743 ± 1.599 |
7.65 ± 1.518 | 7.104 ± 1.522 |
1.776 ± 0.812 | 4.918 ± 1.274 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
