
Solitalea longa
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Solitalea
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
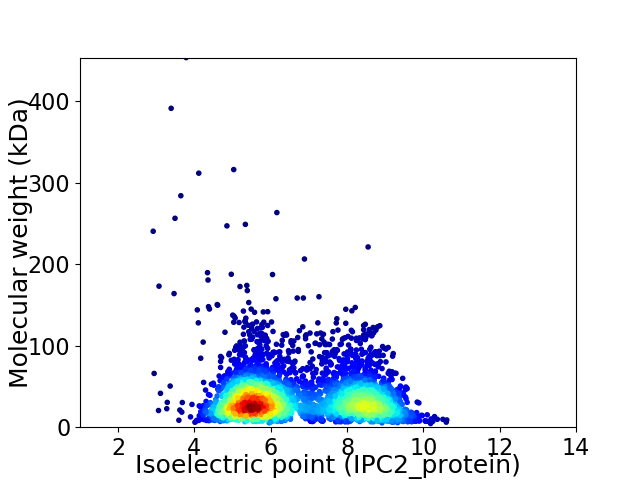
Virtual 2D-PAGE plot for 3780 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S4ZWT7|A0A2S4ZWT7_9SPHI Short-chain dehydrogenase OS=Solitalea longa OX=2079460 GN=C3K47_18705 PE=3 SV=1
YYY2 pKa = 9.99GSDDD6 pKa = 3.06FEEE9 pKa = 4.1QVSDDD14 pKa = 3.42HHH16 pKa = 6.47GTVSQLINITISPVNDDD33 pKa = 3.17PAAADDD39 pKa = 3.22HH40 pKa = 6.85YYY42 pKa = 11.16MDDD45 pKa = 4.15EE46 pKa = 4.99DD47 pKa = 5.96DD48 pKa = 4.1TQTGNVLTDDD58 pKa = 3.78PAASDDD64 pKa = 3.1DDD66 pKa = 4.39DDD68 pKa = 3.96LTAVLINDDD77 pKa = 4.93PPEEE81 pKa = 4.64GTLLLNADDD90 pKa = 4.2SFSFIPAADDD100 pKa = 3.96SGTVSFSYYY109 pKa = 9.66ACDDD113 pKa = 2.92GQPQLCSEEE122 pKa = 4.35EE123 pKa = 4.02SVTITVSPVNDDD135 pKa = 3.53PVATSPVQYYY145 pKa = 9.79EE146 pKa = 4.25NEEE149 pKa = 4.15DD150 pKa = 3.97PVEEE154 pKa = 4.21DD155 pKa = 4.34LVASDDD161 pKa = 4.19DDD163 pKa = 3.89DDD165 pKa = 3.98LTFSKKK171 pKa = 10.93SDDD174 pKa = 3.71ANGTVTVAADDD185 pKa = 3.66SFTYYY190 pKa = 10.68PAANYYY196 pKa = 10.3HH197 pKa = 6.23DDD199 pKa = 3.42DD200 pKa = 3.56FTARR204 pKa = 11.84VSDDD208 pKa = 3.93NGGSVVISVNITVMPVNDDD227 pKa = 4.22PTAVSPISIATDDD240 pKa = 3.2EE241 pKa = 4.15VVFNGQLTADDD252 pKa = 4.16DD253 pKa = 4.54DDD255 pKa = 4.31EEE257 pKa = 4.68LTFTLEEE264 pKa = 4.03QSAHHH269 pKa = 4.75TVVLGNTSHHH279 pKa = 7.02GTTSTLEEE287 pKa = 3.9YY288 pKa = 9.38YYY290 pKa = 10.69PAGGYYY296 pKa = 10.17KKK298 pKa = 9.76DD299 pKa = 3.48DD300 pKa = 3.77HH301 pKa = 6.55QVQIKKK307 pKa = 9.72DD308 pKa = 3.36AGATTLIHHH317 pKa = 5.46HHH319 pKa = 5.47LVNPVNDDD327 pKa = 3.77PVIEEE332 pKa = 4.35AGPFTIDDD340 pKa = 3.72EE341 pKa = 4.38DD342 pKa = 4.46EEE344 pKa = 4.18TGVFKKK350 pKa = 10.6TDDD353 pKa = 3.18DDD355 pKa = 4.22DDD357 pKa = 4.2LTFSVQTAPGTGTLTLQADDD377 pKa = 4.39SFSYYY382 pKa = 10.91PLANYYY388 pKa = 9.68GSDDD392 pKa = 3.21FEEE395 pKa = 4.1QVSDDD400 pKa = 3.42HHH402 pKa = 6.47GTVSQLINITISPVNDDD419 pKa = 3.17PAAADDD425 pKa = 3.22HH426 pKa = 6.85YYY428 pKa = 11.17MDDD431 pKa = 3.76EE432 pKa = 4.98DD433 pKa = 4.97GTQTGNVLTDDD444 pKa = 3.78PAASDDD450 pKa = 3.1DDD452 pKa = 4.39DDD454 pKa = 3.96LTAVLINDDD463 pKa = 4.74PPEEE467 pKa = 4.42EE468 pKa = 4.52TLLLNADDD476 pKa = 4.1SFSFTPAADDD486 pKa = 4.14SGTASFSYYY495 pKa = 9.5ACDDD499 pKa = 2.94GLPQLCSEEE508 pKa = 4.43EE509 pKa = 4.06SVTITVNSVNDDD521 pKa = 3.67PVADDD526 pKa = 4.68SIQYYY531 pKa = 10.03EE532 pKa = 4.48TEEE535 pKa = 4.05DD536 pKa = 3.68PVEEE540 pKa = 4.07DD541 pKa = 3.98LIVTDDD547 pKa = 5.27DD548 pKa = 5.12DD549 pKa = 4.87DDD551 pKa = 3.55LTFVLKKK558 pKa = 10.55GAEEE562 pKa = 4.05QHHH565 pKa = 6.56IVSLTIDDD573 pKa = 3.37AFVYYY578 pKa = 10.73PTANYYY584 pKa = 10.59GTDDD588 pKa = 3.28FTVIVSDDD596 pKa = 3.93QGGNIEEE603 pKa = 4.52IINIQIDDD611 pKa = 4.07VNDDD615 pKa = 3.45PVAVSPIALTTDDD628 pKa = 2.93EE629 pKa = 5.28DD630 pKa = 4.4PVSGQITVSDDD641 pKa = 3.63DDD643 pKa = 4.18DDD645 pKa = 4.04LSYYY649 pKa = 10.77SSEEE653 pKa = 4.26VNGSVIVNPDDD664 pKa = 2.59SFTYYY669 pKa = 9.84PKKK672 pKa = 10.19DDD674 pKa = 3.74YY675 pKa = 9.83GKKK678 pKa = 10.28DD679 pKa = 3.24LVVTITDDD687 pKa = 3.76NGGNAVVKKK696 pKa = 10.0YYY698 pKa = 10.86EEE700 pKa = 4.09NPVNDDD706 pKa = 3.84PVAITPIEEE715 pKa = 4.03QTNKKK720 pKa = 10.51IAVGSNLSATDDD732 pKa = 3.94EEE734 pKa = 5.12EE735 pKa = 4.2DD736 pKa = 3.14LMYYY740 pKa = 10.46VAANPQQGSLIVNADDD756 pKa = 2.87SFIYYY761 pKa = 9.96PNAGFVGVDDD771 pKa = 3.22HH772 pKa = 7.01SAFVDDD778 pKa = 3.99DD779 pKa = 5.01NGGTASIQVNINVLPINNSPRR800 pKa = 11.84ATLIQAFTVNEEE812 pKa = 4.23DD813 pKa = 3.47EE814 pKa = 4.53LSATVEEE821 pKa = 3.99SDDD824 pKa = 4.72DDD826 pKa = 3.84DDD828 pKa = 3.98LNFVILTPASNGQLQLNQQTGAFIYYY854 pKa = 9.91PNKKK858 pKa = 10.04DD859 pKa = 3.7YY860 pKa = 11.62GTDDD864 pKa = 3.18FSVNIDDD871 pKa = 3.74DD872 pKa = 4.61NEEE875 pKa = 3.69LIQVLVNISIIPVNDDD891 pKa = 2.98PIAVTPVSEEE901 pKa = 4.16LEEE904 pKa = 4.26EE905 pKa = 4.73DD906 pKa = 3.63SLSSSVSATDDD917 pKa = 4.46DDD919 pKa = 4.9DD920 pKa = 5.76DD921 pKa = 4.62LTYYY925 pKa = 10.56INTVPLHHH933 pKa = 6.53DDD935 pKa = 3.03LINDDD940 pKa = 4.53DD941 pKa = 3.16TYYY944 pKa = 9.37YYY946 pKa = 11.0PFADDD951 pKa = 4.55NGTDDD956 pKa = 3.54FIVTVDDD963 pKa = 3.7DD964 pKa = 4.63NGGNAQITVQLEEE977 pKa = 3.9TPDDD981 pKa = 3.09DDD983 pKa = 3.48PVAQNISLITDDD995 pKa = 3.26EE996 pKa = 5.95DD997 pKa = 3.72QLTGTLTATDDD1008 pKa = 3.39DDD1010 pKa = 4.22DDD1012 pKa = 3.98LAFTVISEEE1021 pKa = 4.2ANGLFEEE1028 pKa = 5.45SQNGDDD1034 pKa = 3.38YY1035 pKa = 10.81YYY1037 pKa = 10.81PYYY1040 pKa = 10.95NFNGADDD1047 pKa = 3.54VSMQVNDDD1055 pKa = 4.08NGGLASFKKK1064 pKa = 10.74YYY1066 pKa = 10.5EEE1068 pKa = 3.79RR1069 pKa = 11.84SVYYY1073 pKa = 9.95DD1074 pKa = 3.31PIAFEEE1080 pKa = 4.07SFVMDDD1086 pKa = 4.18EE1087 pKa = 4.8EEE1089 pKa = 4.15LNNKKK1094 pKa = 9.69DDD1096 pKa = 4.77ISVDDD1101 pKa = 3.75NPLTFEEE1108 pKa = 5.0ITSTQNGIVQVNADDD1123 pKa = 3.47SFTYYY1128 pKa = 10.12PNPNFNGSDDD1138 pKa = 3.46FKKK1141 pKa = 11.06RR1142 pKa = 11.84VKKK1145 pKa = 10.69DD1146 pKa = 3.87DD1147 pKa = 3.49KK1148 pKa = 11.61GSDDD1152 pKa = 3.92EE1153 pKa = 4.28TVSILINPVNSPPEEE1168 pKa = 3.72ATPINLTINEEE1179 pKa = 4.16DD1180 pKa = 3.2PYYY1183 pKa = 10.36GKKK1186 pKa = 10.4IAVDDD1191 pKa = 3.69EEE1193 pKa = 4.22DDD1195 pKa = 4.06ITFVIGAVVPEEE1207 pKa = 4.68HH1208 pKa = 6.1TLTVNSDDD1216 pKa = 2.73SYYY1219 pKa = 10.75YYY1221 pKa = 10.92PAKKK1225 pKa = 10.41DD1226 pKa = 3.81YY1227 pKa = 10.87GSDDD1231 pKa = 3.14FTARR1235 pKa = 11.84VQDDD1239 pKa = 4.02DD1240 pKa = 3.61KK1241 pKa = 12.0GLSTIAVNITIVPVNDDD1258 pKa = 3.89PVSSNLAYYY1267 pKa = 10.3DD1268 pKa = 3.52LFNEEE1273 pKa = 4.56RR1274 pKa = 11.84TLNISEEE1281 pKa = 4.33VSDDD1285 pKa = 4.49DDD1287 pKa = 3.89PLPLVYYY1294 pKa = 10.22VSTAPQHHH1302 pKa = 5.33TITQMEEE1309 pKa = 4.64GNPVYYY1315 pKa = 10.47PDDD1318 pKa = 4.04NYYY1321 pKa = 10.82GPDDD1325 pKa = 3.13FAYYY1329 pKa = 7.31VCDDD1333 pKa = 3.53FSPEEE1338 pKa = 3.71CTTSTVEEE1346 pKa = 3.7TVKKK1350 pKa = 10.56KK1351 pKa = 10.67LVDDD1355 pKa = 3.93QITKKK1360 pKa = 9.01ASITKKK1366 pKa = 9.8DDD1368 pKa = 4.24NQALSFNLLVKKK1380 pKa = 11.03SDDD1383 pKa = 3.42VNAATQVTVIDDD1395 pKa = 3.97LPEEE1399 pKa = 4.02FSYYY1403 pKa = 11.18SANATLGNVTITGSNIVTWEEE1424 pKa = 3.83SSLAAQQSAQLTIDDD1439 pKa = 3.3TAVQGGDDD1447 pKa = 4.15INSATVTAIEEE1458 pKa = 4.55EEE1460 pKa = 4.45NPSDDD1465 pKa = 3.58KKK1467 pKa = 10.73SVSVEEE1473 pKa = 3.73FGFKKK1478 pKa = 9.74ANVFTPNGDDD1488 pKa = 3.99INDDD1492 pKa = 3.68FTINGVEEE1500 pKa = 4.3DD1501 pKa = 4.33DDD1503 pKa = 6.37EEE1505 pKa = 4.42QVFNRR1510 pKa = 11.84WGAEEE1515 pKa = 3.7YYY1517 pKa = 10.17KK1518 pKa = 10.35KKK1520 pKa = 10.32YYY1522 pKa = 10.62KK1523 pKa = 10.3DDD1525 pKa = 3.09NGSDDD1530 pKa = 4.45PEEE1533 pKa = 3.92TYYY1536 pKa = 10.25YYY1538 pKa = 10.76LRR1540 pKa = 11.84IKKK1543 pKa = 10.66KKK1545 pKa = 9.03GNTKKK1550 pKa = 9.9YYY1552 pKa = 10.1GYYY1555 pKa = 8.6TILRR1559 pKa = 11.84
YYY2 pKa = 9.99GSDDD6 pKa = 3.06FEEE9 pKa = 4.1QVSDDD14 pKa = 3.42HHH16 pKa = 6.47GTVSQLINITISPVNDDD33 pKa = 3.17PAAADDD39 pKa = 3.22HH40 pKa = 6.85YYY42 pKa = 11.16MDDD45 pKa = 4.15EE46 pKa = 4.99DD47 pKa = 5.96DD48 pKa = 4.1TQTGNVLTDDD58 pKa = 3.78PAASDDD64 pKa = 3.1DDD66 pKa = 4.39DDD68 pKa = 3.96LTAVLINDDD77 pKa = 4.93PPEEE81 pKa = 4.64GTLLLNADDD90 pKa = 4.2SFSFIPAADDD100 pKa = 3.96SGTVSFSYYY109 pKa = 9.66ACDDD113 pKa = 2.92GQPQLCSEEE122 pKa = 4.35EE123 pKa = 4.02SVTITVSPVNDDD135 pKa = 3.53PVATSPVQYYY145 pKa = 9.79EE146 pKa = 4.25NEEE149 pKa = 4.15DD150 pKa = 3.97PVEEE154 pKa = 4.21DD155 pKa = 4.34LVASDDD161 pKa = 4.19DDD163 pKa = 3.89DDD165 pKa = 3.98LTFSKKK171 pKa = 10.93SDDD174 pKa = 3.71ANGTVTVAADDD185 pKa = 3.66SFTYYY190 pKa = 10.68PAANYYY196 pKa = 10.3HH197 pKa = 6.23DDD199 pKa = 3.42DD200 pKa = 3.56FTARR204 pKa = 11.84VSDDD208 pKa = 3.93NGGSVVISVNITVMPVNDDD227 pKa = 4.22PTAVSPISIATDDD240 pKa = 3.2EE241 pKa = 4.15VVFNGQLTADDD252 pKa = 4.16DD253 pKa = 4.54DDD255 pKa = 4.31EEE257 pKa = 4.68LTFTLEEE264 pKa = 4.03QSAHHH269 pKa = 4.75TVVLGNTSHHH279 pKa = 7.02GTTSTLEEE287 pKa = 3.9YY288 pKa = 9.38YYY290 pKa = 10.69PAGGYYY296 pKa = 10.17KKK298 pKa = 9.76DD299 pKa = 3.48DD300 pKa = 3.77HH301 pKa = 6.55QVQIKKK307 pKa = 9.72DD308 pKa = 3.36AGATTLIHHH317 pKa = 5.46HHH319 pKa = 5.47LVNPVNDDD327 pKa = 3.77PVIEEE332 pKa = 4.35AGPFTIDDD340 pKa = 3.72EE341 pKa = 4.38DD342 pKa = 4.46EEE344 pKa = 4.18TGVFKKK350 pKa = 10.6TDDD353 pKa = 3.18DDD355 pKa = 4.22DDD357 pKa = 4.2LTFSVQTAPGTGTLTLQADDD377 pKa = 4.39SFSYYY382 pKa = 10.91PLANYYY388 pKa = 9.68GSDDD392 pKa = 3.21FEEE395 pKa = 4.1QVSDDD400 pKa = 3.42HHH402 pKa = 6.47GTVSQLINITISPVNDDD419 pKa = 3.17PAAADDD425 pKa = 3.22HH426 pKa = 6.85YYY428 pKa = 11.17MDDD431 pKa = 3.76EE432 pKa = 4.98DD433 pKa = 4.97GTQTGNVLTDDD444 pKa = 3.78PAASDDD450 pKa = 3.1DDD452 pKa = 4.39DDD454 pKa = 3.96LTAVLINDDD463 pKa = 4.74PPEEE467 pKa = 4.42EE468 pKa = 4.52TLLLNADDD476 pKa = 4.1SFSFTPAADDD486 pKa = 4.14SGTASFSYYY495 pKa = 9.5ACDDD499 pKa = 2.94GLPQLCSEEE508 pKa = 4.43EE509 pKa = 4.06SVTITVNSVNDDD521 pKa = 3.67PVADDD526 pKa = 4.68SIQYYY531 pKa = 10.03EE532 pKa = 4.48TEEE535 pKa = 4.05DD536 pKa = 3.68PVEEE540 pKa = 4.07DD541 pKa = 3.98LIVTDDD547 pKa = 5.27DD548 pKa = 5.12DD549 pKa = 4.87DDD551 pKa = 3.55LTFVLKKK558 pKa = 10.55GAEEE562 pKa = 4.05QHHH565 pKa = 6.56IVSLTIDDD573 pKa = 3.37AFVYYY578 pKa = 10.73PTANYYY584 pKa = 10.59GTDDD588 pKa = 3.28FTVIVSDDD596 pKa = 3.93QGGNIEEE603 pKa = 4.52IINIQIDDD611 pKa = 4.07VNDDD615 pKa = 3.45PVAVSPIALTTDDD628 pKa = 2.93EE629 pKa = 5.28DD630 pKa = 4.4PVSGQITVSDDD641 pKa = 3.63DDD643 pKa = 4.18DDD645 pKa = 4.04LSYYY649 pKa = 10.77SSEEE653 pKa = 4.26VNGSVIVNPDDD664 pKa = 2.59SFTYYY669 pKa = 9.84PKKK672 pKa = 10.19DDD674 pKa = 3.74YY675 pKa = 9.83GKKK678 pKa = 10.28DD679 pKa = 3.24LVVTITDDD687 pKa = 3.76NGGNAVVKKK696 pKa = 10.0YYY698 pKa = 10.86EEE700 pKa = 4.09NPVNDDD706 pKa = 3.84PVAITPIEEE715 pKa = 4.03QTNKKK720 pKa = 10.51IAVGSNLSATDDD732 pKa = 3.94EEE734 pKa = 5.12EE735 pKa = 4.2DD736 pKa = 3.14LMYYY740 pKa = 10.46VAANPQQGSLIVNADDD756 pKa = 2.87SFIYYY761 pKa = 9.96PNAGFVGVDDD771 pKa = 3.22HH772 pKa = 7.01SAFVDDD778 pKa = 3.99DD779 pKa = 5.01NGGTASIQVNINVLPINNSPRR800 pKa = 11.84ATLIQAFTVNEEE812 pKa = 4.23DD813 pKa = 3.47EE814 pKa = 4.53LSATVEEE821 pKa = 3.99SDDD824 pKa = 4.72DDD826 pKa = 3.84DDD828 pKa = 3.98LNFVILTPASNGQLQLNQQTGAFIYYY854 pKa = 9.91PNKKK858 pKa = 10.04DD859 pKa = 3.7YY860 pKa = 11.62GTDDD864 pKa = 3.18FSVNIDDD871 pKa = 3.74DD872 pKa = 4.61NEEE875 pKa = 3.69LIQVLVNISIIPVNDDD891 pKa = 2.98PIAVTPVSEEE901 pKa = 4.16LEEE904 pKa = 4.26EE905 pKa = 4.73DD906 pKa = 3.63SLSSSVSATDDD917 pKa = 4.46DDD919 pKa = 4.9DD920 pKa = 5.76DD921 pKa = 4.62LTYYY925 pKa = 10.56INTVPLHHH933 pKa = 6.53DDD935 pKa = 3.03LINDDD940 pKa = 4.53DD941 pKa = 3.16TYYY944 pKa = 9.37YYY946 pKa = 11.0PFADDD951 pKa = 4.55NGTDDD956 pKa = 3.54FIVTVDDD963 pKa = 3.7DD964 pKa = 4.63NGGNAQITVQLEEE977 pKa = 3.9TPDDD981 pKa = 3.09DDD983 pKa = 3.48PVAQNISLITDDD995 pKa = 3.26EE996 pKa = 5.95DD997 pKa = 3.72QLTGTLTATDDD1008 pKa = 3.39DDD1010 pKa = 4.22DDD1012 pKa = 3.98LAFTVISEEE1021 pKa = 4.2ANGLFEEE1028 pKa = 5.45SQNGDDD1034 pKa = 3.38YY1035 pKa = 10.81YYY1037 pKa = 10.81PYYY1040 pKa = 10.95NFNGADDD1047 pKa = 3.54VSMQVNDDD1055 pKa = 4.08NGGLASFKKK1064 pKa = 10.74YYY1066 pKa = 10.5EEE1068 pKa = 3.79RR1069 pKa = 11.84SVYYY1073 pKa = 9.95DD1074 pKa = 3.31PIAFEEE1080 pKa = 4.07SFVMDDD1086 pKa = 4.18EE1087 pKa = 4.8EEE1089 pKa = 4.15LNNKKK1094 pKa = 9.69DDD1096 pKa = 4.77ISVDDD1101 pKa = 3.75NPLTFEEE1108 pKa = 5.0ITSTQNGIVQVNADDD1123 pKa = 3.47SFTYYY1128 pKa = 10.12PNPNFNGSDDD1138 pKa = 3.46FKKK1141 pKa = 11.06RR1142 pKa = 11.84VKKK1145 pKa = 10.69DD1146 pKa = 3.87DD1147 pKa = 3.49KK1148 pKa = 11.61GSDDD1152 pKa = 3.92EE1153 pKa = 4.28TVSILINPVNSPPEEE1168 pKa = 3.72ATPINLTINEEE1179 pKa = 4.16DD1180 pKa = 3.2PYYY1183 pKa = 10.36GKKK1186 pKa = 10.4IAVDDD1191 pKa = 3.69EEE1193 pKa = 4.22DDD1195 pKa = 4.06ITFVIGAVVPEEE1207 pKa = 4.68HH1208 pKa = 6.1TLTVNSDDD1216 pKa = 2.73SYYY1219 pKa = 10.75YYY1221 pKa = 10.92PAKKK1225 pKa = 10.41DD1226 pKa = 3.81YY1227 pKa = 10.87GSDDD1231 pKa = 3.14FTARR1235 pKa = 11.84VQDDD1239 pKa = 4.02DD1240 pKa = 3.61KK1241 pKa = 12.0GLSTIAVNITIVPVNDDD1258 pKa = 3.89PVSSNLAYYY1267 pKa = 10.3DD1268 pKa = 3.52LFNEEE1273 pKa = 4.56RR1274 pKa = 11.84TLNISEEE1281 pKa = 4.33VSDDD1285 pKa = 4.49DDD1287 pKa = 3.89PLPLVYYY1294 pKa = 10.22VSTAPQHHH1302 pKa = 5.33TITQMEEE1309 pKa = 4.64GNPVYYY1315 pKa = 10.47PDDD1318 pKa = 4.04NYYY1321 pKa = 10.82GPDDD1325 pKa = 3.13FAYYY1329 pKa = 7.31VCDDD1333 pKa = 3.53FSPEEE1338 pKa = 3.71CTTSTVEEE1346 pKa = 3.7TVKKK1350 pKa = 10.56KK1351 pKa = 10.67LVDDD1355 pKa = 3.93QITKKK1360 pKa = 9.01ASITKKK1366 pKa = 9.8DDD1368 pKa = 4.24NQALSFNLLVKKK1380 pKa = 11.03SDDD1383 pKa = 3.42VNAATQVTVIDDD1395 pKa = 3.97LPEEE1399 pKa = 4.02FSYYY1403 pKa = 11.18SANATLGNVTITGSNIVTWEEE1424 pKa = 3.83SSLAAQQSAQLTIDDD1439 pKa = 3.3TAVQGGDDD1447 pKa = 4.15INSATVTAIEEE1458 pKa = 4.55EEE1460 pKa = 4.45NPSDDD1465 pKa = 3.58KKK1467 pKa = 10.73SVSVEEE1473 pKa = 3.73FGFKKK1478 pKa = 9.74ANVFTPNGDDD1488 pKa = 3.99INDDD1492 pKa = 3.68FTINGVEEE1500 pKa = 4.3DD1501 pKa = 4.33DDD1503 pKa = 6.37EEE1505 pKa = 4.42QVFNRR1510 pKa = 11.84WGAEEE1515 pKa = 3.7YYY1517 pKa = 10.17KK1518 pKa = 10.35KKK1520 pKa = 10.32YYY1522 pKa = 10.62KK1523 pKa = 10.3DDD1525 pKa = 3.09NGSDDD1530 pKa = 4.45PEEE1533 pKa = 3.92TYYY1536 pKa = 10.25YYY1538 pKa = 10.76LRR1540 pKa = 11.84IKKK1543 pKa = 10.66KKK1545 pKa = 9.03GNTKKK1550 pKa = 9.9YYY1552 pKa = 10.1GYYY1555 pKa = 8.6TILRR1559 pKa = 11.84
Molecular weight: 164.05 kDa
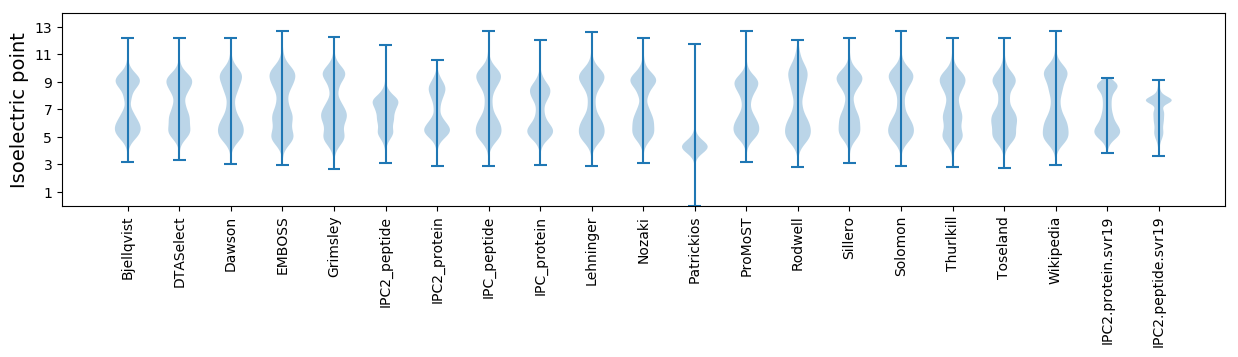
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S4ZZV3|A0A2S4ZZV3_9SPHI Uncharacterized protein OS=Solitalea longa OX=2079460 GN=C3K47_15700 PE=4 SV=1
MM1 pKa = 7.32NNARR5 pKa = 11.84KK6 pKa = 10.01NEE8 pKa = 3.97IKK10 pKa = 10.86DD11 pKa = 3.37NAFKK15 pKa = 11.13VFALVCTSVGLLVLGLLLYY34 pKa = 10.67NIISVGLTRR43 pKa = 11.84INWDD47 pKa = 4.44FITNLPSRR55 pKa = 11.84RR56 pKa = 11.84PGKK59 pKa = 10.43AGIYY63 pKa = 7.46TALLGMIWVVTLTLVISFPIGIGAGVYY90 pKa = 10.25LEE92 pKa = 4.81EE93 pKa = 4.94YY94 pKa = 9.08GAKK97 pKa = 10.02NRR99 pKa = 11.84FAKK102 pKa = 10.51FIEE105 pKa = 4.04INIANLAGVPSIIYY119 pKa = 9.93GILGLEE125 pKa = 3.93LFGRR129 pKa = 11.84LLSLGNSILAASLTLSLLVLPIVIVATRR157 pKa = 11.84EE158 pKa = 4.09AVKK161 pKa = 10.5SVPNSLRR168 pKa = 11.84EE169 pKa = 3.88ASIGLGATRR178 pKa = 11.84WQTISRR184 pKa = 11.84IVLPASFGSILTGLILAVSRR204 pKa = 11.84AVGEE208 pKa = 4.12TAPLIVVGALVYY220 pKa = 11.0VPFAPKK226 pKa = 10.48SPLDD230 pKa = 3.51QFTVLPIQIFNWTSRR245 pKa = 11.84PQSGFVVNAAAAIIILLLFVFILNGIAVFLRR276 pKa = 11.84NRR278 pKa = 11.84HH279 pKa = 4.37NRR281 pKa = 11.84RR282 pKa = 11.84VKK284 pKa = 10.12YY285 pKa = 10.45
MM1 pKa = 7.32NNARR5 pKa = 11.84KK6 pKa = 10.01NEE8 pKa = 3.97IKK10 pKa = 10.86DD11 pKa = 3.37NAFKK15 pKa = 11.13VFALVCTSVGLLVLGLLLYY34 pKa = 10.67NIISVGLTRR43 pKa = 11.84INWDD47 pKa = 4.44FITNLPSRR55 pKa = 11.84RR56 pKa = 11.84PGKK59 pKa = 10.43AGIYY63 pKa = 7.46TALLGMIWVVTLTLVISFPIGIGAGVYY90 pKa = 10.25LEE92 pKa = 4.81EE93 pKa = 4.94YY94 pKa = 9.08GAKK97 pKa = 10.02NRR99 pKa = 11.84FAKK102 pKa = 10.51FIEE105 pKa = 4.04INIANLAGVPSIIYY119 pKa = 9.93GILGLEE125 pKa = 3.93LFGRR129 pKa = 11.84LLSLGNSILAASLTLSLLVLPIVIVATRR157 pKa = 11.84EE158 pKa = 4.09AVKK161 pKa = 10.5SVPNSLRR168 pKa = 11.84EE169 pKa = 3.88ASIGLGATRR178 pKa = 11.84WQTISRR184 pKa = 11.84IVLPASFGSILTGLILAVSRR204 pKa = 11.84AVGEE208 pKa = 4.12TAPLIVVGALVYY220 pKa = 11.0VPFAPKK226 pKa = 10.48SPLDD230 pKa = 3.51QFTVLPIQIFNWTSRR245 pKa = 11.84PQSGFVVNAAAAIIILLLFVFILNGIAVFLRR276 pKa = 11.84NRR278 pKa = 11.84HH279 pKa = 4.37NRR281 pKa = 11.84RR282 pKa = 11.84VKK284 pKa = 10.12YY285 pKa = 10.45
Molecular weight: 30.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322964 |
38 |
4393 |
350.0 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.086 ± 0.041 | 0.859 ± 0.015 |
5.222 ± 0.029 | 6.015 ± 0.048 |
5.081 ± 0.03 | 6.627 ± 0.043 |
1.726 ± 0.017 | 7.41 ± 0.037 |
7.34 ± 0.051 | 9.418 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.223 ± 0.021 | 5.953 ± 0.035 |
3.681 ± 0.026 | 3.645 ± 0.022 |
3.527 ± 0.026 | 6.688 ± 0.037 |
5.87 ± 0.064 | 6.462 ± 0.034 |
1.125 ± 0.015 | 4.041 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
