
Valsa malicola
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Diaporthales; Valsaceae; Valsa
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
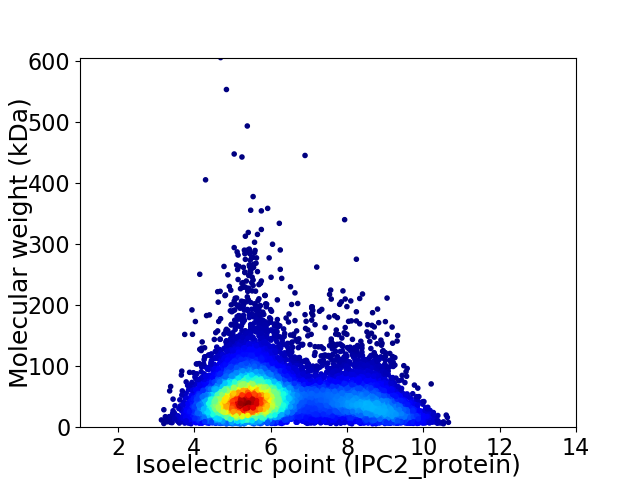
Virtual 2D-PAGE plot for 10560 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
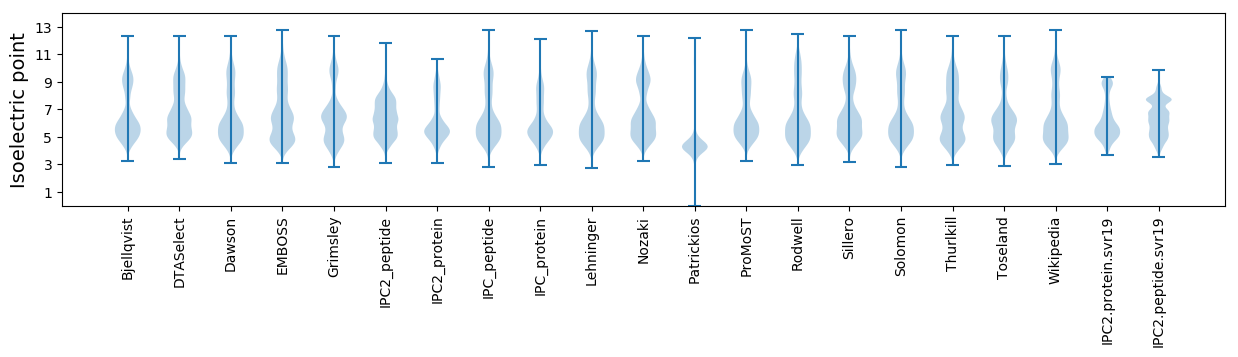
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A423X6L7|A0A423X6L7_9PEZI Mediator of RNA polymerase II transcription subunit 16 OS=Valsa malicola OX=356882 GN=MED16 PE=3 SV=1
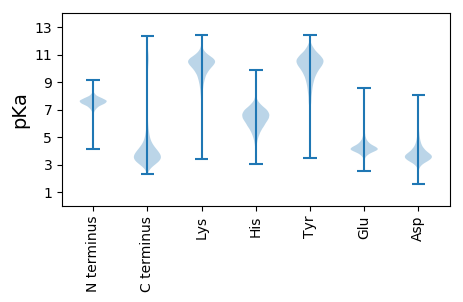
MM1 pKa = 7.36HH2 pKa = 8.34LIRR5 pKa = 11.84PVWAVGTALLLASSSTAQTSTNCDD29 pKa = 3.47PTQTTCDD36 pKa = 3.75PDD38 pKa = 3.45EE39 pKa = 4.54ALGMAINVDD48 pKa = 4.59FTQGEE53 pKa = 4.69VNSFTSSGGTPSYY66 pKa = 11.14GSDD69 pKa = 2.71GVTFTISGSGDD80 pKa = 3.3APQLNSMFYY89 pKa = 10.58IMFGKK94 pKa = 10.26VDD96 pKa = 3.38ITMKK100 pKa = 10.09VAPGTGIVTSVVLQSDD116 pKa = 4.11DD117 pKa = 3.97LDD119 pKa = 4.73EE120 pKa = 6.67IDD122 pKa = 4.93MEE124 pKa = 4.71FLGSDD129 pKa = 3.55DD130 pKa = 4.94SKK132 pKa = 11.04VQLMYY137 pKa = 10.61FGKK140 pKa = 10.59GDD142 pKa = 3.59RR143 pKa = 11.84ADD145 pKa = 3.67NAVVIADD152 pKa = 4.05APDD155 pKa = 3.55NQSNFVTYY163 pKa = 10.02TIDD166 pKa = 3.13WSSDD170 pKa = 3.0RR171 pKa = 11.84IAWSVGGTTVRR182 pKa = 11.84VLEE185 pKa = 4.3STTYY189 pKa = 10.02PDD191 pKa = 4.76YY192 pKa = 11.32YY193 pKa = 9.89PQTPMQLKK201 pKa = 9.38FGIWAGGDD209 pKa = 3.32SGNTQGTIDD218 pKa = 3.6WAGGATDD225 pKa = 3.96YY226 pKa = 11.63SSGPFSAVVQSVAVTDD242 pKa = 3.91YY243 pKa = 10.55STGSSYY249 pKa = 11.02TYY251 pKa = 11.03GDD253 pKa = 3.5TSGTWGSIEE262 pKa = 4.12SDD264 pKa = 2.96GGEE267 pKa = 4.06VNANAGEE274 pKa = 4.2AATVTSVSGATSTDD288 pKa = 2.79SSNFPGSGIGDD299 pKa = 3.61GTSTVVASVNSMPSGWHH316 pKa = 4.25MTSNGKK322 pKa = 8.89IVPNSSTVVKK332 pKa = 9.73PPHH335 pKa = 6.09VLLLAGPLSFFIFVLCNGIWRR356 pKa = 4.18
MM1 pKa = 7.36HH2 pKa = 8.34LIRR5 pKa = 11.84PVWAVGTALLLASSSTAQTSTNCDD29 pKa = 3.47PTQTTCDD36 pKa = 3.75PDD38 pKa = 3.45EE39 pKa = 4.54ALGMAINVDD48 pKa = 4.59FTQGEE53 pKa = 4.69VNSFTSSGGTPSYY66 pKa = 11.14GSDD69 pKa = 2.71GVTFTISGSGDD80 pKa = 3.3APQLNSMFYY89 pKa = 10.58IMFGKK94 pKa = 10.26VDD96 pKa = 3.38ITMKK100 pKa = 10.09VAPGTGIVTSVVLQSDD116 pKa = 4.11DD117 pKa = 3.97LDD119 pKa = 4.73EE120 pKa = 6.67IDD122 pKa = 4.93MEE124 pKa = 4.71FLGSDD129 pKa = 3.55DD130 pKa = 4.94SKK132 pKa = 11.04VQLMYY137 pKa = 10.61FGKK140 pKa = 10.59GDD142 pKa = 3.59RR143 pKa = 11.84ADD145 pKa = 3.67NAVVIADD152 pKa = 4.05APDD155 pKa = 3.55NQSNFVTYY163 pKa = 10.02TIDD166 pKa = 3.13WSSDD170 pKa = 3.0RR171 pKa = 11.84IAWSVGGTTVRR182 pKa = 11.84VLEE185 pKa = 4.3STTYY189 pKa = 10.02PDD191 pKa = 4.76YY192 pKa = 11.32YY193 pKa = 9.89PQTPMQLKK201 pKa = 9.38FGIWAGGDD209 pKa = 3.32SGNTQGTIDD218 pKa = 3.6WAGGATDD225 pKa = 3.96YY226 pKa = 11.63SSGPFSAVVQSVAVTDD242 pKa = 3.91YY243 pKa = 10.55STGSSYY249 pKa = 11.02TYY251 pKa = 11.03GDD253 pKa = 3.5TSGTWGSIEE262 pKa = 4.12SDD264 pKa = 2.96GGEE267 pKa = 4.06VNANAGEE274 pKa = 4.2AATVTSVSGATSTDD288 pKa = 2.79SSNFPGSGIGDD299 pKa = 3.61GTSTVVASVNSMPSGWHH316 pKa = 4.25MTSNGKK322 pKa = 8.89IVPNSSTVVKK332 pKa = 9.73PPHH335 pKa = 6.09VLLLAGPLSFFIFVLCNGIWRR356 pKa = 4.18
Molecular weight: 37.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A423VVT6|A0A423VVT6_9PEZI MFS domain-containing protein OS=Valsa malicola OX=356882 GN=VMCG_08552 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.17RR3 pKa = 11.84TRR5 pKa = 11.84RR6 pKa = 11.84LEE8 pKa = 4.29DD9 pKa = 3.18DD10 pKa = 3.78TAVLMVPIYY19 pKa = 9.97DD20 pKa = 3.97EE21 pKa = 4.36RR22 pKa = 11.84QQGADD27 pKa = 2.99LLMFPVSDD35 pKa = 4.59EE36 pKa = 4.1SRR38 pKa = 11.84QPQQPQQPQPQPQSQPPQQLPQKK61 pKa = 10.4LPQQQQTPPQQPLPQYY77 pKa = 9.3QSHH80 pKa = 5.63VTPQQRR86 pKa = 11.84QQPQQPQRR94 pKa = 11.84IEE96 pKa = 4.14TPTARR101 pKa = 11.84PPTPPPKK108 pKa = 9.75PQLPLEE114 pKa = 4.49CEE116 pKa = 4.01DD117 pKa = 3.36WDD119 pKa = 5.35DD120 pKa = 3.85YY121 pKa = 12.03LMWRR125 pKa = 11.84RR126 pKa = 11.84AAEE129 pKa = 4.05AASLSDD135 pKa = 3.27QQQQQQHH142 pKa = 5.95GPSSSSRR149 pKa = 11.84WRR151 pKa = 11.84RR152 pKa = 11.84RR153 pKa = 11.84ILHH156 pKa = 5.54QHH158 pKa = 5.47QQHH161 pKa = 5.68QPPTAPKK168 pKa = 9.85EE169 pKa = 4.3DD170 pKa = 3.33ILRR173 pKa = 11.84GFEE176 pKa = 3.69EE177 pKa = 4.04WKK179 pKa = 10.39EE180 pKa = 3.68YY181 pKa = 10.59LAASRR186 pKa = 11.84AASRR190 pKa = 11.84RR191 pKa = 11.84GGRR194 pKa = 11.84PISIISTSTTTTTTGRR210 pKa = 11.84SFKK213 pKa = 10.18RR214 pKa = 11.84SSMCSISSTKK224 pKa = 10.14SGKK227 pKa = 8.83PRR229 pKa = 11.84SLRR232 pKa = 11.84RR233 pKa = 11.84CSRR236 pKa = 11.84SRR238 pKa = 11.84HH239 pKa = 4.16LRR241 pKa = 11.84AYY243 pKa = 9.72YY244 pKa = 10.42SEE246 pKa = 5.01GSLSTARR253 pKa = 11.84NNQRR257 pKa = 11.84VDD259 pKa = 3.47GPLKK263 pKa = 8.93EE264 pKa = 4.45TIVNVDD270 pKa = 3.21HH271 pKa = 6.66VPRR274 pKa = 11.84GRR276 pKa = 11.84PQFQPRR282 pKa = 11.84QPTSHH287 pKa = 6.12QSLRR291 pKa = 11.84ASWVSEE297 pKa = 4.09SCGPPPHH304 pKa = 7.16PPPDD308 pKa = 3.85KK309 pKa = 10.19PLPALPPIIDD319 pKa = 4.01SPVIDD324 pKa = 4.86ASPIVDD330 pKa = 3.88SFPVEE335 pKa = 3.69RR336 pKa = 11.84RR337 pKa = 11.84AKK339 pKa = 9.83RR340 pKa = 11.84RR341 pKa = 11.84NRR343 pKa = 11.84SLLDD347 pKa = 3.07EE348 pKa = 4.52RR349 pKa = 11.84EE350 pKa = 3.84RR351 pKa = 11.84KK352 pKa = 9.52RR353 pKa = 11.84FHH355 pKa = 6.2RR356 pKa = 11.84VYY358 pKa = 9.67WHH360 pKa = 6.6
MM1 pKa = 7.28KK2 pKa = 9.17RR3 pKa = 11.84TRR5 pKa = 11.84RR6 pKa = 11.84LEE8 pKa = 4.29DD9 pKa = 3.18DD10 pKa = 3.78TAVLMVPIYY19 pKa = 9.97DD20 pKa = 3.97EE21 pKa = 4.36RR22 pKa = 11.84QQGADD27 pKa = 2.99LLMFPVSDD35 pKa = 4.59EE36 pKa = 4.1SRR38 pKa = 11.84QPQQPQQPQPQPQSQPPQQLPQKK61 pKa = 10.4LPQQQQTPPQQPLPQYY77 pKa = 9.3QSHH80 pKa = 5.63VTPQQRR86 pKa = 11.84QQPQQPQRR94 pKa = 11.84IEE96 pKa = 4.14TPTARR101 pKa = 11.84PPTPPPKK108 pKa = 9.75PQLPLEE114 pKa = 4.49CEE116 pKa = 4.01DD117 pKa = 3.36WDD119 pKa = 5.35DD120 pKa = 3.85YY121 pKa = 12.03LMWRR125 pKa = 11.84RR126 pKa = 11.84AAEE129 pKa = 4.05AASLSDD135 pKa = 3.27QQQQQQHH142 pKa = 5.95GPSSSSRR149 pKa = 11.84WRR151 pKa = 11.84RR152 pKa = 11.84RR153 pKa = 11.84ILHH156 pKa = 5.54QHH158 pKa = 5.47QQHH161 pKa = 5.68QPPTAPKK168 pKa = 9.85EE169 pKa = 4.3DD170 pKa = 3.33ILRR173 pKa = 11.84GFEE176 pKa = 3.69EE177 pKa = 4.04WKK179 pKa = 10.39EE180 pKa = 3.68YY181 pKa = 10.59LAASRR186 pKa = 11.84AASRR190 pKa = 11.84RR191 pKa = 11.84GGRR194 pKa = 11.84PISIISTSTTTTTTGRR210 pKa = 11.84SFKK213 pKa = 10.18RR214 pKa = 11.84SSMCSISSTKK224 pKa = 10.14SGKK227 pKa = 8.83PRR229 pKa = 11.84SLRR232 pKa = 11.84RR233 pKa = 11.84CSRR236 pKa = 11.84SRR238 pKa = 11.84HH239 pKa = 4.16LRR241 pKa = 11.84AYY243 pKa = 9.72YY244 pKa = 10.42SEE246 pKa = 5.01GSLSTARR253 pKa = 11.84NNQRR257 pKa = 11.84VDD259 pKa = 3.47GPLKK263 pKa = 8.93EE264 pKa = 4.45TIVNVDD270 pKa = 3.21HH271 pKa = 6.66VPRR274 pKa = 11.84GRR276 pKa = 11.84PQFQPRR282 pKa = 11.84QPTSHH287 pKa = 6.12QSLRR291 pKa = 11.84ASWVSEE297 pKa = 4.09SCGPPPHH304 pKa = 7.16PPPDD308 pKa = 3.85KK309 pKa = 10.19PLPALPPIIDD319 pKa = 4.01SPVIDD324 pKa = 4.86ASPIVDD330 pKa = 3.88SFPVEE335 pKa = 3.69RR336 pKa = 11.84RR337 pKa = 11.84AKK339 pKa = 9.83RR340 pKa = 11.84RR341 pKa = 11.84NRR343 pKa = 11.84SLLDD347 pKa = 3.07EE348 pKa = 4.52RR349 pKa = 11.84EE350 pKa = 3.84RR351 pKa = 11.84KK352 pKa = 9.52RR353 pKa = 11.84FHH355 pKa = 6.2RR356 pKa = 11.84VYY358 pKa = 9.67WHH360 pKa = 6.6
Molecular weight: 41.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5240010 |
50 |
5565 |
496.2 |
54.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.905 ± 0.02 | 1.142 ± 0.007 |
6.009 ± 0.016 | 6.305 ± 0.023 |
3.545 ± 0.014 | 7.388 ± 0.021 |
2.352 ± 0.011 | 4.554 ± 0.014 |
4.752 ± 0.022 | 8.601 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.286 ± 0.009 | 3.58 ± 0.012 |
6.117 ± 0.026 | 3.96 ± 0.019 |
6.057 ± 0.022 | 7.96 ± 0.027 |
6.0 ± 0.02 | 6.286 ± 0.017 |
1.47 ± 0.009 | 2.729 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
