
Human associated huchismacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Huchismacovirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
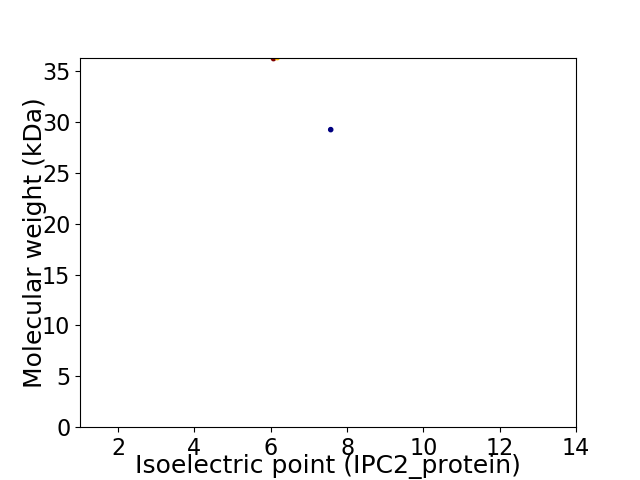
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5GNX7|A0A0B5GNX7_9VIRU Rep protein OS=Human associated huchismacovirus 2 OX=2169935 GN=rep PE=4 SV=1
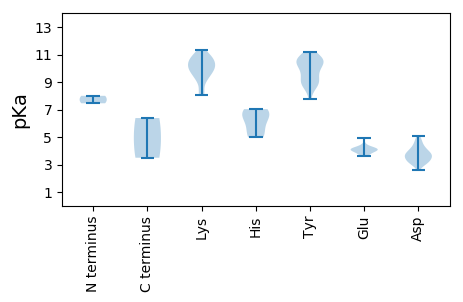
MM1 pKa = 7.46TNFVSQKK8 pKa = 9.09FQWFVDD14 pKa = 4.15LQTSSDD20 pKa = 3.41SMQVVTITAGGQRR33 pKa = 11.84VRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84LLPFFAAFKK46 pKa = 9.4YY47 pKa = 10.09YY48 pKa = 10.8KK49 pKa = 9.96LGSVSVRR56 pKa = 11.84FVPASTLPVDD66 pKa = 3.92PTGLSYY72 pKa = 10.98SAGEE76 pKa = 4.1NTVDD80 pKa = 5.06PRR82 pKa = 11.84DD83 pKa = 3.46QFNPGLVRR91 pKa = 11.84ITNGEE96 pKa = 4.11DD97 pKa = 3.45FNIDD101 pKa = 3.29TALLADD107 pKa = 4.25DD108 pKa = 4.15TASQNMYY115 pKa = 8.68YY116 pKa = 10.67QLMLDD121 pKa = 3.58PRR123 pKa = 11.84WYY125 pKa = 10.63KK126 pKa = 10.84FNLQSGFRR134 pKa = 11.84RR135 pKa = 11.84KK136 pKa = 9.91AIPLYY141 pKa = 9.2WSIGQLHH148 pKa = 6.14QDD150 pKa = 3.67VFPGAVQNLPSVTKK164 pKa = 10.46RR165 pKa = 11.84SSGSVNSYY173 pKa = 8.94DD174 pKa = 3.08TGNRR178 pKa = 11.84IDD180 pKa = 4.42HH181 pKa = 6.78IEE183 pKa = 4.2NGSAAFNFYY192 pKa = 10.58SGSNPHH198 pKa = 7.03SIFQVGGKK206 pKa = 9.91DD207 pKa = 3.04RR208 pKa = 11.84MGWMPTDD215 pKa = 3.58AFYY218 pKa = 11.18DD219 pKa = 3.75PGNPDD224 pKa = 3.2LTLGSMAQVPEE235 pKa = 4.19VEE237 pKa = 4.48LLRR240 pKa = 11.84VILPKK245 pKa = 10.27AYY247 pKa = 7.79KK248 pKa = 9.62TKK250 pKa = 10.4FFYY253 pKa = 10.52RR254 pKa = 11.84LYY256 pKa = 10.4IEE258 pKa = 3.94EE259 pKa = 4.25TVNFKK264 pKa = 11.31GPVSMYY270 pKa = 9.15ATNSSVAGSFYY281 pKa = 10.56PLNQVDD287 pKa = 3.78RR288 pKa = 11.84FIYY291 pKa = 9.9PVMNEE296 pKa = 4.33LGSTTQAPARR306 pKa = 11.84NPINGSDD313 pKa = 3.08WHH315 pKa = 6.67NDD317 pKa = 2.62GNAGGEE323 pKa = 4.1NN324 pKa = 3.5
MM1 pKa = 7.46TNFVSQKK8 pKa = 9.09FQWFVDD14 pKa = 4.15LQTSSDD20 pKa = 3.41SMQVVTITAGGQRR33 pKa = 11.84VRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84LLPFFAAFKK46 pKa = 9.4YY47 pKa = 10.09YY48 pKa = 10.8KK49 pKa = 9.96LGSVSVRR56 pKa = 11.84FVPASTLPVDD66 pKa = 3.92PTGLSYY72 pKa = 10.98SAGEE76 pKa = 4.1NTVDD80 pKa = 5.06PRR82 pKa = 11.84DD83 pKa = 3.46QFNPGLVRR91 pKa = 11.84ITNGEE96 pKa = 4.11DD97 pKa = 3.45FNIDD101 pKa = 3.29TALLADD107 pKa = 4.25DD108 pKa = 4.15TASQNMYY115 pKa = 8.68YY116 pKa = 10.67QLMLDD121 pKa = 3.58PRR123 pKa = 11.84WYY125 pKa = 10.63KK126 pKa = 10.84FNLQSGFRR134 pKa = 11.84RR135 pKa = 11.84KK136 pKa = 9.91AIPLYY141 pKa = 9.2WSIGQLHH148 pKa = 6.14QDD150 pKa = 3.67VFPGAVQNLPSVTKK164 pKa = 10.46RR165 pKa = 11.84SSGSVNSYY173 pKa = 8.94DD174 pKa = 3.08TGNRR178 pKa = 11.84IDD180 pKa = 4.42HH181 pKa = 6.78IEE183 pKa = 4.2NGSAAFNFYY192 pKa = 10.58SGSNPHH198 pKa = 7.03SIFQVGGKK206 pKa = 9.91DD207 pKa = 3.04RR208 pKa = 11.84MGWMPTDD215 pKa = 3.58AFYY218 pKa = 11.18DD219 pKa = 3.75PGNPDD224 pKa = 3.2LTLGSMAQVPEE235 pKa = 4.19VEE237 pKa = 4.48LLRR240 pKa = 11.84VILPKK245 pKa = 10.27AYY247 pKa = 7.79KK248 pKa = 9.62TKK250 pKa = 10.4FFYY253 pKa = 10.52RR254 pKa = 11.84LYY256 pKa = 10.4IEE258 pKa = 3.94EE259 pKa = 4.25TVNFKK264 pKa = 11.31GPVSMYY270 pKa = 9.15ATNSSVAGSFYY281 pKa = 10.56PLNQVDD287 pKa = 3.78RR288 pKa = 11.84FIYY291 pKa = 9.9PVMNEE296 pKa = 4.33LGSTTQAPARR306 pKa = 11.84NPINGSDD313 pKa = 3.08WHH315 pKa = 6.67NDD317 pKa = 2.62GNAGGEE323 pKa = 4.1NN324 pKa = 3.5
Molecular weight: 36.23 kDa
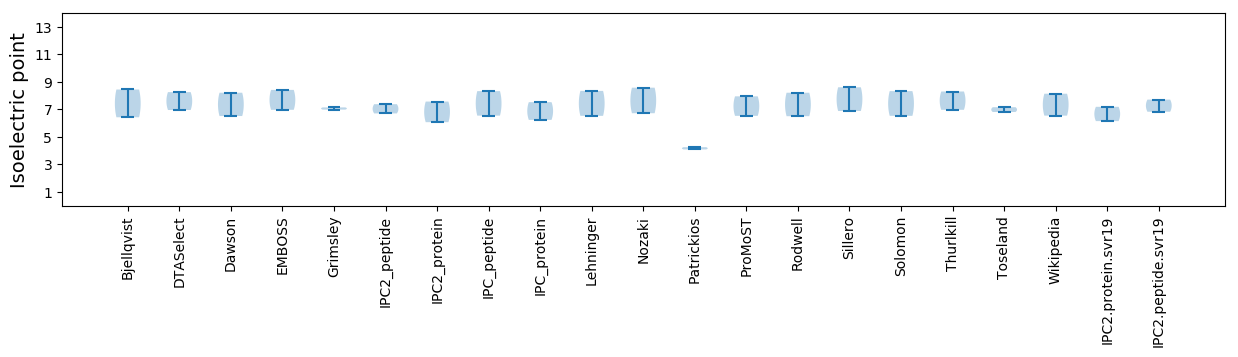
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5GNX7|A0A0B5GNX7_9VIRU Rep protein OS=Human associated huchismacovirus 2 OX=2169935 GN=rep PE=4 SV=1
MM1 pKa = 8.0PSPKK5 pKa = 9.58WYY7 pKa = 9.89DD8 pKa = 3.07ITVSADD14 pKa = 3.23KK15 pKa = 11.1VPEE18 pKa = 3.86KK19 pKa = 10.7TMVEE23 pKa = 4.13WLNKK27 pKa = 8.11YY28 pKa = 8.88CEE30 pKa = 4.0RR31 pKa = 11.84YY32 pKa = 9.93AYY34 pKa = 10.21GRR36 pKa = 11.84EE37 pKa = 4.0TGEE40 pKa = 3.82NGYY43 pKa = 9.55KK44 pKa = 10.03HH45 pKa = 4.98YY46 pKa = 10.62QIRR49 pKa = 11.84LVLKK53 pKa = 8.29TGADD57 pKa = 3.1ISEE60 pKa = 4.08MRR62 pKa = 11.84KK63 pKa = 8.03VWSAFGHH70 pKa = 5.33VSCTSVRR77 pKa = 11.84NFDD80 pKa = 3.83YY81 pKa = 10.89VLKK84 pKa = 10.68EE85 pKa = 3.62GDD87 pKa = 4.61YY88 pKa = 8.56VCSWIKK94 pKa = 10.52IPEE97 pKa = 4.0NMKK100 pKa = 10.64NPEE103 pKa = 3.63FRR105 pKa = 11.84PWQRR109 pKa = 11.84ALLDD113 pKa = 3.89LEE115 pKa = 4.19QDD117 pKa = 3.5DD118 pKa = 4.25RR119 pKa = 11.84QIDD122 pKa = 4.11VIIDD126 pKa = 3.5LQGNSGKK133 pKa = 10.16SWLTKK138 pKa = 9.84YY139 pKa = 10.33CALKK143 pKa = 9.74MRR145 pKa = 11.84AINVPSGLEE154 pKa = 3.86GKK156 pKa = 10.17DD157 pKa = 3.27IMRR160 pKa = 11.84MCLKK164 pKa = 10.52RR165 pKa = 11.84GAQSGLYY172 pKa = 9.5IFDD175 pKa = 3.76MPRR178 pKa = 11.84AITKK182 pKa = 9.11QSKK185 pKa = 9.26SVWSAIEE192 pKa = 4.23SIKK195 pKa = 10.93NGYY198 pKa = 8.87LWDD201 pKa = 4.71DD202 pKa = 3.86RR203 pKa = 11.84YY204 pKa = 8.77TWEE207 pKa = 4.92EE208 pKa = 3.62KK209 pKa = 10.09WIDD212 pKa = 3.66PPRR215 pKa = 11.84IWVFTNEE222 pKa = 3.93YY223 pKa = 9.29PKK225 pKa = 10.9YY226 pKa = 10.45DD227 pKa = 3.78LVSEE231 pKa = 4.11DD232 pKa = 4.79RR233 pKa = 11.84LRR235 pKa = 11.84FWAPTPTGLVSLAHH249 pKa = 6.38
MM1 pKa = 8.0PSPKK5 pKa = 9.58WYY7 pKa = 9.89DD8 pKa = 3.07ITVSADD14 pKa = 3.23KK15 pKa = 11.1VPEE18 pKa = 3.86KK19 pKa = 10.7TMVEE23 pKa = 4.13WLNKK27 pKa = 8.11YY28 pKa = 8.88CEE30 pKa = 4.0RR31 pKa = 11.84YY32 pKa = 9.93AYY34 pKa = 10.21GRR36 pKa = 11.84EE37 pKa = 4.0TGEE40 pKa = 3.82NGYY43 pKa = 9.55KK44 pKa = 10.03HH45 pKa = 4.98YY46 pKa = 10.62QIRR49 pKa = 11.84LVLKK53 pKa = 8.29TGADD57 pKa = 3.1ISEE60 pKa = 4.08MRR62 pKa = 11.84KK63 pKa = 8.03VWSAFGHH70 pKa = 5.33VSCTSVRR77 pKa = 11.84NFDD80 pKa = 3.83YY81 pKa = 10.89VLKK84 pKa = 10.68EE85 pKa = 3.62GDD87 pKa = 4.61YY88 pKa = 8.56VCSWIKK94 pKa = 10.52IPEE97 pKa = 4.0NMKK100 pKa = 10.64NPEE103 pKa = 3.63FRR105 pKa = 11.84PWQRR109 pKa = 11.84ALLDD113 pKa = 3.89LEE115 pKa = 4.19QDD117 pKa = 3.5DD118 pKa = 4.25RR119 pKa = 11.84QIDD122 pKa = 4.11VIIDD126 pKa = 3.5LQGNSGKK133 pKa = 10.16SWLTKK138 pKa = 9.84YY139 pKa = 10.33CALKK143 pKa = 9.74MRR145 pKa = 11.84AINVPSGLEE154 pKa = 3.86GKK156 pKa = 10.17DD157 pKa = 3.27IMRR160 pKa = 11.84MCLKK164 pKa = 10.52RR165 pKa = 11.84GAQSGLYY172 pKa = 9.5IFDD175 pKa = 3.76MPRR178 pKa = 11.84AITKK182 pKa = 9.11QSKK185 pKa = 9.26SVWSAIEE192 pKa = 4.23SIKK195 pKa = 10.93NGYY198 pKa = 8.87LWDD201 pKa = 4.71DD202 pKa = 3.86RR203 pKa = 11.84YY204 pKa = 8.77TWEE207 pKa = 4.92EE208 pKa = 3.62KK209 pKa = 10.09WIDD212 pKa = 3.66PPRR215 pKa = 11.84IWVFTNEE222 pKa = 3.93YY223 pKa = 9.29PKK225 pKa = 10.9YY226 pKa = 10.45DD227 pKa = 3.78LVSEE231 pKa = 4.11DD232 pKa = 4.79RR233 pKa = 11.84LRR235 pKa = 11.84FWAPTPTGLVSLAHH249 pKa = 6.38
Molecular weight: 29.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
573 |
249 |
324 |
286.5 |
32.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.585 ± 0.472 | 0.873 ± 0.701 |
6.457 ± 0.228 | 4.363 ± 1.273 |
4.712 ± 1.421 | 6.981 ± 0.839 |
1.222 ± 0.01 | 4.887 ± 0.95 |
5.41 ± 1.619 | 7.155 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.967 ± 0.152 | 5.585 ± 1.216 |
5.934 ± 0.44 | 4.014 ± 0.742 |
5.759 ± 0.411 | 7.853 ± 0.633 |
5.236 ± 0.505 | 6.806 ± 0.483 |
2.967 ± 1.144 | 5.236 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
