
Beihai picobirna-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
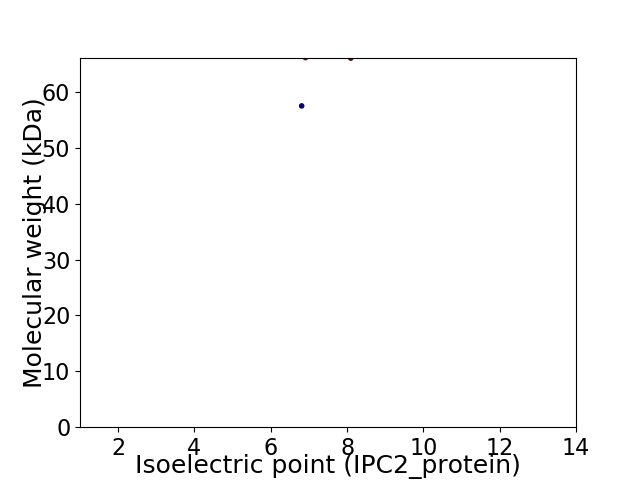
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLI7|A0A1L3KLI7_9VIRU Uncharacterized protein OS=Beihai picobirna-like virus 8 OX=1922525 PE=4 SV=1
MM1 pKa = 7.24VKK3 pKa = 10.3HH4 pKa = 5.76NQFSFLDD11 pKa = 3.78SLDD14 pKa = 3.43LDD16 pKa = 4.04KK17 pKa = 11.07EE18 pKa = 4.5VKK20 pKa = 10.16LRR22 pKa = 11.84LSSNLLNIIQGNDD35 pKa = 3.03TVLYY39 pKa = 9.24SPLVKK44 pKa = 10.13SVKK47 pKa = 9.86PEE49 pKa = 4.34SILTGWDD56 pKa = 3.48KK57 pKa = 10.93IFKK60 pKa = 9.93SKK62 pKa = 9.71SVKK65 pKa = 8.95MNKK68 pKa = 8.98VLHH71 pKa = 6.65DD72 pKa = 4.41LEE74 pKa = 6.56DD75 pKa = 4.64DD76 pKa = 3.63NRR78 pKa = 11.84SKK80 pKa = 11.19YY81 pKa = 10.23GPRR84 pKa = 11.84SIAVPWVDD92 pKa = 3.12RR93 pKa = 11.84RR94 pKa = 11.84SDD96 pKa = 3.4VLSYY100 pKa = 11.0FEE102 pKa = 4.64SNNMEE107 pKa = 4.41RR108 pKa = 11.84PPLGLPSLRR117 pKa = 11.84RR118 pKa = 11.84NLRR121 pKa = 11.84PLEE124 pKa = 3.98LDD126 pKa = 3.51KK127 pKa = 11.26AIKK130 pKa = 9.98LLKK133 pKa = 10.28NDD135 pKa = 4.03TNSGLPYY142 pKa = 9.83YY143 pKa = 9.95RR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 10.71GGLKK150 pKa = 10.21DD151 pKa = 3.24IYY153 pKa = 10.89LKK155 pKa = 10.78EE156 pKa = 4.03FTNILPRR163 pKa = 11.84EE164 pKa = 4.51DD165 pKa = 3.59PCVMFTRR172 pKa = 11.84TQEE175 pKa = 4.04GKK177 pKa = 8.45KK178 pKa = 8.45TRR180 pKa = 11.84TVWGYY185 pKa = 10.41PMADD189 pKa = 3.6TVNEE193 pKa = 4.09MCVYY197 pKa = 10.56SPLLGYY203 pKa = 7.94QRR205 pKa = 11.84KK206 pKa = 9.51LNWRR210 pKa = 11.84SCLNSPDD217 pKa = 3.52AVNIEE222 pKa = 3.88ITKK225 pKa = 10.86LIDD228 pKa = 3.2KK229 pKa = 10.2ALKK232 pKa = 10.02QDD234 pKa = 3.42VSLLSIDD241 pKa = 4.28FSAYY245 pKa = 9.93DD246 pKa = 3.44ASLDD250 pKa = 4.24RR251 pKa = 11.84PLQKK255 pKa = 10.7AAFEE259 pKa = 4.56YY260 pKa = 10.17ISKK263 pKa = 10.18LFQPSYY269 pKa = 11.39EE270 pKa = 3.96EE271 pKa = 5.24LINNLFNRR279 pKa = 11.84FNTIGLVTPDD289 pKa = 4.68GILSGHH295 pKa = 6.68HH296 pKa = 5.99GVPSGSTFTNEE307 pKa = 3.13VDD309 pKa = 5.39SIIQYY314 pKa = 10.63LIALDD319 pKa = 4.05YY320 pKa = 11.05GLEE323 pKa = 4.24DD324 pKa = 4.27SSMQIQGDD332 pKa = 3.76DD333 pKa = 3.18GVYY336 pKa = 10.17CVQDD340 pKa = 3.42GTALAEE346 pKa = 4.15HH347 pKa = 6.7FKK349 pKa = 11.35SFGLNVNFEE358 pKa = 3.81KK359 pKa = 10.49SYY361 pKa = 10.61YY362 pKa = 9.85SKK364 pKa = 10.77DD365 pKa = 3.39YY366 pKa = 10.93CVYY369 pKa = 10.62LQNLYY374 pKa = 10.88HH375 pKa = 6.81NDD377 pKa = 3.34YY378 pKa = 9.97RR379 pKa = 11.84DD380 pKa = 3.55SSGKK384 pKa = 9.56ICGIYY389 pKa = 7.99PTYY392 pKa = 10.42RR393 pKa = 11.84ALNRR397 pKa = 11.84LIHH400 pKa = 6.14PEE402 pKa = 3.91RR403 pKa = 11.84FVDD406 pKa = 5.13FEE408 pKa = 5.14DD409 pKa = 3.54IKK411 pKa = 11.13ISGRR415 pKa = 11.84DD416 pKa = 3.43YY417 pKa = 10.78FSIRR421 pKa = 11.84SICILEE427 pKa = 3.92NCKK430 pKa = 10.1YY431 pKa = 10.5HH432 pKa = 7.29PLFEE436 pKa = 5.56DD437 pKa = 3.82LVKK440 pKa = 10.91YY441 pKa = 10.37IYY443 pKa = 10.94KK444 pKa = 10.01LDD446 pKa = 3.67KK447 pKa = 11.24YY448 pKa = 9.82NLKK451 pKa = 10.44FSRR454 pKa = 11.84DD455 pKa = 3.26SLFRR459 pKa = 11.84FVQNRR464 pKa = 11.84KK465 pKa = 8.92QEE467 pKa = 4.18SGVQGIFNYY476 pKa = 10.18RR477 pKa = 11.84YY478 pKa = 8.66EE479 pKa = 5.33DD480 pKa = 3.97DD481 pKa = 3.65VSGIDD486 pKa = 3.24SFEE489 pKa = 4.12TMKK492 pKa = 11.0VLNKK496 pKa = 10.39LL497 pKa = 3.46
MM1 pKa = 7.24VKK3 pKa = 10.3HH4 pKa = 5.76NQFSFLDD11 pKa = 3.78SLDD14 pKa = 3.43LDD16 pKa = 4.04KK17 pKa = 11.07EE18 pKa = 4.5VKK20 pKa = 10.16LRR22 pKa = 11.84LSSNLLNIIQGNDD35 pKa = 3.03TVLYY39 pKa = 9.24SPLVKK44 pKa = 10.13SVKK47 pKa = 9.86PEE49 pKa = 4.34SILTGWDD56 pKa = 3.48KK57 pKa = 10.93IFKK60 pKa = 9.93SKK62 pKa = 9.71SVKK65 pKa = 8.95MNKK68 pKa = 8.98VLHH71 pKa = 6.65DD72 pKa = 4.41LEE74 pKa = 6.56DD75 pKa = 4.64DD76 pKa = 3.63NRR78 pKa = 11.84SKK80 pKa = 11.19YY81 pKa = 10.23GPRR84 pKa = 11.84SIAVPWVDD92 pKa = 3.12RR93 pKa = 11.84RR94 pKa = 11.84SDD96 pKa = 3.4VLSYY100 pKa = 11.0FEE102 pKa = 4.64SNNMEE107 pKa = 4.41RR108 pKa = 11.84PPLGLPSLRR117 pKa = 11.84RR118 pKa = 11.84NLRR121 pKa = 11.84PLEE124 pKa = 3.98LDD126 pKa = 3.51KK127 pKa = 11.26AIKK130 pKa = 9.98LLKK133 pKa = 10.28NDD135 pKa = 4.03TNSGLPYY142 pKa = 9.83YY143 pKa = 9.95RR144 pKa = 11.84RR145 pKa = 11.84KK146 pKa = 10.71GGLKK150 pKa = 10.21DD151 pKa = 3.24IYY153 pKa = 10.89LKK155 pKa = 10.78EE156 pKa = 4.03FTNILPRR163 pKa = 11.84EE164 pKa = 4.51DD165 pKa = 3.59PCVMFTRR172 pKa = 11.84TQEE175 pKa = 4.04GKK177 pKa = 8.45KK178 pKa = 8.45TRR180 pKa = 11.84TVWGYY185 pKa = 10.41PMADD189 pKa = 3.6TVNEE193 pKa = 4.09MCVYY197 pKa = 10.56SPLLGYY203 pKa = 7.94QRR205 pKa = 11.84KK206 pKa = 9.51LNWRR210 pKa = 11.84SCLNSPDD217 pKa = 3.52AVNIEE222 pKa = 3.88ITKK225 pKa = 10.86LIDD228 pKa = 3.2KK229 pKa = 10.2ALKK232 pKa = 10.02QDD234 pKa = 3.42VSLLSIDD241 pKa = 4.28FSAYY245 pKa = 9.93DD246 pKa = 3.44ASLDD250 pKa = 4.24RR251 pKa = 11.84PLQKK255 pKa = 10.7AAFEE259 pKa = 4.56YY260 pKa = 10.17ISKK263 pKa = 10.18LFQPSYY269 pKa = 11.39EE270 pKa = 3.96EE271 pKa = 5.24LINNLFNRR279 pKa = 11.84FNTIGLVTPDD289 pKa = 4.68GILSGHH295 pKa = 6.68HH296 pKa = 5.99GVPSGSTFTNEE307 pKa = 3.13VDD309 pKa = 5.39SIIQYY314 pKa = 10.63LIALDD319 pKa = 4.05YY320 pKa = 11.05GLEE323 pKa = 4.24DD324 pKa = 4.27SSMQIQGDD332 pKa = 3.76DD333 pKa = 3.18GVYY336 pKa = 10.17CVQDD340 pKa = 3.42GTALAEE346 pKa = 4.15HH347 pKa = 6.7FKK349 pKa = 11.35SFGLNVNFEE358 pKa = 3.81KK359 pKa = 10.49SYY361 pKa = 10.61YY362 pKa = 9.85SKK364 pKa = 10.77DD365 pKa = 3.39YY366 pKa = 10.93CVYY369 pKa = 10.62LQNLYY374 pKa = 10.88HH375 pKa = 6.81NDD377 pKa = 3.34YY378 pKa = 9.97RR379 pKa = 11.84DD380 pKa = 3.55SSGKK384 pKa = 9.56ICGIYY389 pKa = 7.99PTYY392 pKa = 10.42RR393 pKa = 11.84ALNRR397 pKa = 11.84LIHH400 pKa = 6.14PEE402 pKa = 3.91RR403 pKa = 11.84FVDD406 pKa = 5.13FEE408 pKa = 5.14DD409 pKa = 3.54IKK411 pKa = 11.13ISGRR415 pKa = 11.84DD416 pKa = 3.43YY417 pKa = 10.78FSIRR421 pKa = 11.84SICILEE427 pKa = 3.92NCKK430 pKa = 10.1YY431 pKa = 10.5HH432 pKa = 7.29PLFEE436 pKa = 5.56DD437 pKa = 3.82LVKK440 pKa = 10.91YY441 pKa = 10.37IYY443 pKa = 10.94KK444 pKa = 10.01LDD446 pKa = 3.67KK447 pKa = 11.24YY448 pKa = 9.82NLKK451 pKa = 10.44FSRR454 pKa = 11.84DD455 pKa = 3.26SLFRR459 pKa = 11.84FVQNRR464 pKa = 11.84KK465 pKa = 8.92QEE467 pKa = 4.18SGVQGIFNYY476 pKa = 10.18RR477 pKa = 11.84YY478 pKa = 8.66EE479 pKa = 5.33DD480 pKa = 3.97DD481 pKa = 3.65VSGIDD486 pKa = 3.24SFEE489 pKa = 4.12TMKK492 pKa = 11.0VLNKK496 pKa = 10.39LL497 pKa = 3.46
Molecular weight: 57.53 kDa
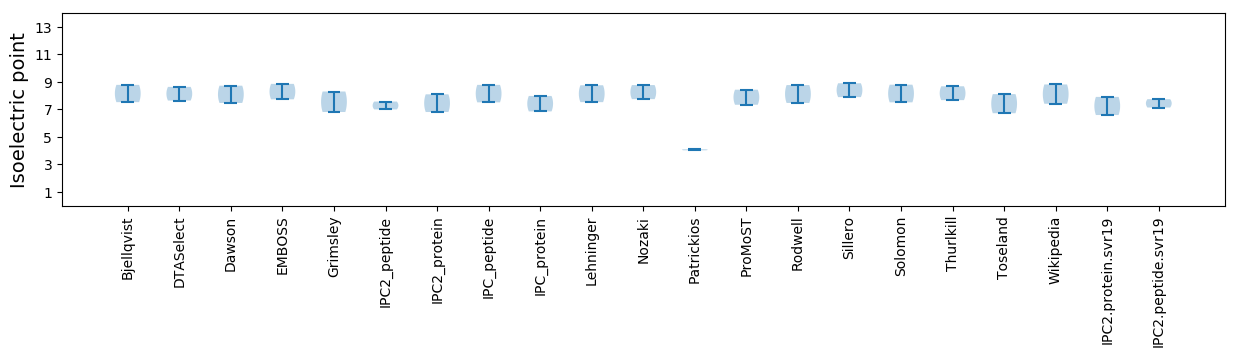
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLI7|A0A1L3KLI7_9VIRU Uncharacterized protein OS=Beihai picobirna-like virus 8 OX=1922525 PE=4 SV=1
MM1 pKa = 6.37YY2 pKa = 8.89TSNFIYY8 pKa = 9.87LKK10 pKa = 10.83HH11 pKa = 6.25MNNEE15 pKa = 3.75YY16 pKa = 10.61KK17 pKa = 10.7KK18 pKa = 10.63PFFRR22 pKa = 11.84KK23 pKa = 9.87NKK25 pKa = 7.13TDD27 pKa = 3.43KK28 pKa = 11.43NIDD31 pKa = 3.44SDD33 pKa = 4.45SSVSTDD39 pKa = 2.71STQRR43 pKa = 11.84NEE45 pKa = 4.45DD46 pKa = 3.68STSSLQASGNPAFVPGGARR65 pKa = 11.84PWGVTGSQSIPPMGVSRR82 pKa = 11.84NVYY85 pKa = 8.53RR86 pKa = 11.84QINGGVARR94 pKa = 11.84ALGSDD99 pKa = 2.82IGKK102 pKa = 10.16AVIGEE107 pKa = 4.16VASYY111 pKa = 11.46GLTKK115 pKa = 10.78LNSKK119 pKa = 10.75LNDD122 pKa = 3.49FEE124 pKa = 6.32SEE126 pKa = 4.2FEE128 pKa = 4.05NRR130 pKa = 11.84KK131 pKa = 8.76PPRR134 pKa = 11.84NKK136 pKa = 10.44GKK138 pKa = 8.6GTRR141 pKa = 11.84KK142 pKa = 9.65RR143 pKa = 11.84GRR145 pKa = 11.84GNRR148 pKa = 11.84DD149 pKa = 2.98SGSGNGGGGSDD160 pKa = 3.66NNYY163 pKa = 10.24SGNSSGEE170 pKa = 3.62RR171 pKa = 11.84GVPYY175 pKa = 10.17GGSPMPYY182 pKa = 8.19RR183 pKa = 11.84NRR185 pKa = 11.84YY186 pKa = 8.78ILDD189 pKa = 3.48TGMSASTIIDD199 pKa = 3.94DD200 pKa = 4.64DD201 pKa = 4.88QNTTDD206 pKa = 3.37HH207 pKa = 7.07WSPLLTTNGSLLDD220 pKa = 3.96ILWVPQTRR228 pKa = 11.84TYY230 pKa = 11.44DD231 pKa = 3.47VIRR234 pKa = 11.84TYY236 pKa = 11.37VFDD239 pKa = 4.54KK240 pKa = 11.31YY241 pKa = 11.06LDD243 pKa = 3.89KK244 pKa = 11.06LQQDD248 pKa = 4.77LNPEE252 pKa = 4.03YY253 pKa = 10.7TIDD256 pKa = 3.47TDD258 pKa = 3.93VFGSYY263 pKa = 10.66LSDD266 pKa = 3.19VSEE269 pKa = 4.51ALQAYY274 pKa = 6.94YY275 pKa = 10.49TLGHH279 pKa = 5.99IRR281 pKa = 11.84AFAFGPYY288 pKa = 8.87RR289 pKa = 11.84QQNIALYY296 pKa = 10.1KK297 pKa = 8.96IGRR300 pKa = 11.84RR301 pKa = 11.84YY302 pKa = 10.47GSDD305 pKa = 3.17VIDD308 pKa = 3.15AHH310 pKa = 7.12RR311 pKa = 11.84YY312 pKa = 8.75LKK314 pKa = 10.81SKK316 pKa = 10.92LEE318 pKa = 4.03NLPIPPRR325 pKa = 11.84LVHH328 pKa = 6.07FMYY331 pKa = 10.88YY332 pKa = 8.68FTQNFTMHH340 pKa = 6.95PTPASPCIRR349 pKa = 11.84INYY352 pKa = 9.62RR353 pKa = 11.84GMFLPGKK360 pKa = 7.97NRR362 pKa = 11.84KK363 pKa = 7.88WKK365 pKa = 10.34DD366 pKa = 3.25YY367 pKa = 10.73GVNCSISADD376 pKa = 3.79LFKK379 pKa = 11.39GLIDD383 pKa = 3.97NLDD386 pKa = 3.6KK387 pKa = 11.28HH388 pKa = 6.61RR389 pKa = 11.84LTYY392 pKa = 11.04NKK394 pKa = 9.51INRR397 pKa = 11.84ICPEE401 pKa = 4.18LKK403 pKa = 10.15VGQLMDD409 pKa = 4.51PSCEE413 pKa = 4.12VIYY416 pKa = 10.56DD417 pKa = 3.82EE418 pKa = 6.01QFNTWWHH425 pKa = 4.95NHH427 pKa = 4.85CTSYY431 pKa = 10.61WEE433 pKa = 4.57GGKK436 pKa = 8.08LTYY439 pKa = 9.08TRR441 pKa = 11.84NVADD445 pKa = 4.4LRR447 pKa = 11.84LTAYY451 pKa = 10.07YY452 pKa = 10.15GQYY455 pKa = 9.91TNRR458 pKa = 11.84LDD460 pKa = 3.56TAIYY464 pKa = 10.11ASSDD468 pKa = 3.41LAVRR472 pKa = 11.84NAEE475 pKa = 3.78GDD477 pKa = 3.7YY478 pKa = 11.26YY479 pKa = 10.3MYY481 pKa = 10.57PGVWCPFNGWKK492 pKa = 8.27NTDD495 pKa = 3.19AVKK498 pKa = 10.89DD499 pKa = 4.01EE500 pKa = 4.89GPLTSINYY508 pKa = 7.79WRR510 pKa = 11.84EE511 pKa = 3.6DD512 pKa = 2.88KK513 pKa = 10.51IYY515 pKa = 10.74NMNYY519 pKa = 8.91TSSKK523 pKa = 10.92EE524 pKa = 3.98NVKK527 pKa = 9.64WGPFSNVYY535 pKa = 9.65KK536 pKa = 10.96VPLIDD541 pKa = 4.39DD542 pKa = 3.88HH543 pKa = 6.46EE544 pKa = 4.59KK545 pKa = 10.4KK546 pKa = 10.52IKK548 pKa = 10.37IFTGSDD554 pKa = 3.17CGVQIAQEE562 pKa = 4.02HH563 pKa = 5.76NAALMGDD570 pKa = 3.72QAKK573 pKa = 10.65KK574 pKa = 9.43LISWIFEE581 pKa = 4.07TT582 pKa = 5.44
MM1 pKa = 6.37YY2 pKa = 8.89TSNFIYY8 pKa = 9.87LKK10 pKa = 10.83HH11 pKa = 6.25MNNEE15 pKa = 3.75YY16 pKa = 10.61KK17 pKa = 10.7KK18 pKa = 10.63PFFRR22 pKa = 11.84KK23 pKa = 9.87NKK25 pKa = 7.13TDD27 pKa = 3.43KK28 pKa = 11.43NIDD31 pKa = 3.44SDD33 pKa = 4.45SSVSTDD39 pKa = 2.71STQRR43 pKa = 11.84NEE45 pKa = 4.45DD46 pKa = 3.68STSSLQASGNPAFVPGGARR65 pKa = 11.84PWGVTGSQSIPPMGVSRR82 pKa = 11.84NVYY85 pKa = 8.53RR86 pKa = 11.84QINGGVARR94 pKa = 11.84ALGSDD99 pKa = 2.82IGKK102 pKa = 10.16AVIGEE107 pKa = 4.16VASYY111 pKa = 11.46GLTKK115 pKa = 10.78LNSKK119 pKa = 10.75LNDD122 pKa = 3.49FEE124 pKa = 6.32SEE126 pKa = 4.2FEE128 pKa = 4.05NRR130 pKa = 11.84KK131 pKa = 8.76PPRR134 pKa = 11.84NKK136 pKa = 10.44GKK138 pKa = 8.6GTRR141 pKa = 11.84KK142 pKa = 9.65RR143 pKa = 11.84GRR145 pKa = 11.84GNRR148 pKa = 11.84DD149 pKa = 2.98SGSGNGGGGSDD160 pKa = 3.66NNYY163 pKa = 10.24SGNSSGEE170 pKa = 3.62RR171 pKa = 11.84GVPYY175 pKa = 10.17GGSPMPYY182 pKa = 8.19RR183 pKa = 11.84NRR185 pKa = 11.84YY186 pKa = 8.78ILDD189 pKa = 3.48TGMSASTIIDD199 pKa = 3.94DD200 pKa = 4.64DD201 pKa = 4.88QNTTDD206 pKa = 3.37HH207 pKa = 7.07WSPLLTTNGSLLDD220 pKa = 3.96ILWVPQTRR228 pKa = 11.84TYY230 pKa = 11.44DD231 pKa = 3.47VIRR234 pKa = 11.84TYY236 pKa = 11.37VFDD239 pKa = 4.54KK240 pKa = 11.31YY241 pKa = 11.06LDD243 pKa = 3.89KK244 pKa = 11.06LQQDD248 pKa = 4.77LNPEE252 pKa = 4.03YY253 pKa = 10.7TIDD256 pKa = 3.47TDD258 pKa = 3.93VFGSYY263 pKa = 10.66LSDD266 pKa = 3.19VSEE269 pKa = 4.51ALQAYY274 pKa = 6.94YY275 pKa = 10.49TLGHH279 pKa = 5.99IRR281 pKa = 11.84AFAFGPYY288 pKa = 8.87RR289 pKa = 11.84QQNIALYY296 pKa = 10.1KK297 pKa = 8.96IGRR300 pKa = 11.84RR301 pKa = 11.84YY302 pKa = 10.47GSDD305 pKa = 3.17VIDD308 pKa = 3.15AHH310 pKa = 7.12RR311 pKa = 11.84YY312 pKa = 8.75LKK314 pKa = 10.81SKK316 pKa = 10.92LEE318 pKa = 4.03NLPIPPRR325 pKa = 11.84LVHH328 pKa = 6.07FMYY331 pKa = 10.88YY332 pKa = 8.68FTQNFTMHH340 pKa = 6.95PTPASPCIRR349 pKa = 11.84INYY352 pKa = 9.62RR353 pKa = 11.84GMFLPGKK360 pKa = 7.97NRR362 pKa = 11.84KK363 pKa = 7.88WKK365 pKa = 10.34DD366 pKa = 3.25YY367 pKa = 10.73GVNCSISADD376 pKa = 3.79LFKK379 pKa = 11.39GLIDD383 pKa = 3.97NLDD386 pKa = 3.6KK387 pKa = 11.28HH388 pKa = 6.61RR389 pKa = 11.84LTYY392 pKa = 11.04NKK394 pKa = 9.51INRR397 pKa = 11.84ICPEE401 pKa = 4.18LKK403 pKa = 10.15VGQLMDD409 pKa = 4.51PSCEE413 pKa = 4.12VIYY416 pKa = 10.56DD417 pKa = 3.82EE418 pKa = 6.01QFNTWWHH425 pKa = 4.95NHH427 pKa = 4.85CTSYY431 pKa = 10.61WEE433 pKa = 4.57GGKK436 pKa = 8.08LTYY439 pKa = 9.08TRR441 pKa = 11.84NVADD445 pKa = 4.4LRR447 pKa = 11.84LTAYY451 pKa = 10.07YY452 pKa = 10.15GQYY455 pKa = 9.91TNRR458 pKa = 11.84LDD460 pKa = 3.56TAIYY464 pKa = 10.11ASSDD468 pKa = 3.41LAVRR472 pKa = 11.84NAEE475 pKa = 3.78GDD477 pKa = 3.7YY478 pKa = 11.26YY479 pKa = 10.3MYY481 pKa = 10.57PGVWCPFNGWKK492 pKa = 8.27NTDD495 pKa = 3.19AVKK498 pKa = 10.89DD499 pKa = 4.01EE500 pKa = 4.89GPLTSINYY508 pKa = 7.79WRR510 pKa = 11.84EE511 pKa = 3.6DD512 pKa = 2.88KK513 pKa = 10.51IYY515 pKa = 10.74NMNYY519 pKa = 8.91TSSKK523 pKa = 10.92EE524 pKa = 3.98NVKK527 pKa = 9.64WGPFSNVYY535 pKa = 9.65KK536 pKa = 10.96VPLIDD541 pKa = 4.39DD542 pKa = 3.88HH543 pKa = 6.46EE544 pKa = 4.59KK545 pKa = 10.4KK546 pKa = 10.52IKK548 pKa = 10.37IFTGSDD554 pKa = 3.17CGVQIAQEE562 pKa = 4.02HH563 pKa = 5.76NAALMGDD570 pKa = 3.72QAKK573 pKa = 10.65KK574 pKa = 9.43LISWIFEE581 pKa = 4.07TT582 pKa = 5.44
Molecular weight: 66.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1079 |
497 |
582 |
539.5 |
61.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.707 ± 0.738 | 1.39 ± 0.148 |
7.507 ± 0.366 | 4.263 ± 0.518 |
4.078 ± 0.508 | 7.044 ± 1.225 |
1.761 ± 0.102 | 5.931 ± 0.207 |
6.951 ± 0.47 | 8.897 ± 1.738 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.854 ± 0.165 | 6.951 ± 0.482 |
4.634 ± 0.276 | 3.058 ± 0.027 |
5.653 ± 0.013 | 8.341 ± 0.346 |
4.912 ± 1.008 | 5.19 ± 0.436 |
1.483 ± 0.458 | 6.395 ± 0.242 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
